
-
Tim Devarenne
- Associate Head for Undergraduate Programs, Associate Professor, Biochemistry and Biophysics
- Focus Area: Algal hydrocarbons, Botryococcus braunii, plant pathogen interactions, programmed cell death, protein kinase signaling
- Office:
- NMR N218A
- Email:
- timothy.devarenne@ag.tamu.edu
- Phone:
- 979-314-8288
Education
- Undergraduate Education
- B.S. 1991
- Graduate Education
- M.S. 1993
- Ph.D. 2000
- Postdoc. 2001-2006
Areas of Expertise
- Algal hydrocarbons
- Botryococcus braunii
- Plant pathogen interactions
- Programmed cell death
- Protein kinase signaling
Professional Summary
The Devarenne lab focuses on identifying the genes and enzymes involved in hydrocarbon biosynthesis in the green microalga Botryococcus braunii and analyzing how plants control programmed cell death to control resistance to bacterial pathogens.
Algal hydrocarbon biosynthesis
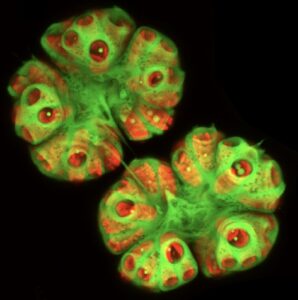
A confocal microscopy image of a B. braunii colony showing chloroplast localization from chlorophyll autofluorescence (red) and stained with Nile red (green) to visualize the liquid hydrocarbons.
Botryococcus braunii is a colony-forming green microalga that is well known for producing large quantities of liquid hydrocarbons, which can be converted into petroleum-equivalent combustion engine fuels. Thus, B. braunii hydrocarbons are a potential renewable source of transportation fuels. Interestingly, B. braunii hydrocarbons have been shown to be a major constituent of currently used petroleum deposits. We are studying the genes involved in the biosynthesis of the B. braunii liquid hydrocarbons from three different chemical races of the alga. Each race produces a different molecular type of hydrocarbon. Our studies include using transcriptomic and genomic data to identify hydrocarbon biosynthesis genes, cloning these genes, and functionally characterizing the enzymes encoded by these genes.
Control of plant programmed cell death
We study the biochemical and molecular mechanisms underlying the control of programmed cell death (PCD) in plants and how PCD is manipulated during plant-pathogen interactions. Specifically, we study the interaction between tomato and Pseudomonas syringae pv. tomato (Pst) the causative agent of bacterial speck disease. During the tomato resistance response to Pst, PCD is induced by the plant to limit the spread of the pathogen. Many of the genes that control plant PCD are serine/threonine (S/T) protein kinase. We are interested in studying a specific class of S/T protein kinases that control PCD in plants called AGC kinases and how they are regulated during the plant resistance response.
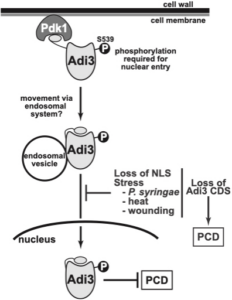
In tomato, the AGC kinase Adi3 is phosphorylated by Pdk1 to direct nuclear localization for control of programmed cell death (PCD) through cell death suppression (CDS). The presence of stresses such as P. syringae prevents Adi3 nuclear localization leading to PCD induction and resistance to the pathogen.
Publications
- View publications on Google Scholar