
-
Paul Straight
- Associate Professor, Biochemistry and Biophysics, Genetics
- Focus Area: bacteria, microbiology, metabolism, antibiotics, signaling, microbial communities, secondary metabolites, bacillus subtilis, streptomyces, motility, bacterial genetics, mass spectrometry
- Office:
- BICH 435A
- Email:
- [email protected]
- Phone:
- 979-314-8293 Lab phone: 979-314-8298
Education
- Undergraduate Education
- B.S. Lewis & Clark College
- Graduate Education
- Ph.D. U. of Colorado, Boulder
- Postdoc. Harvard Medical School
Areas of Expertise
- Microbiology
- Bacterial Genetics
- Natural Products
- Actinomycetes
- Biofilms
- Electron Microscopy
Professional Summary
The Straight lab studies chemical and genetic mechanisms of interaction between species of bacteria. Interspecies interactions are the foundation of the bacterial communities that shape the environment and human health. Recognizing the importance of these microorganisms in the biosphere, there remains much to learn about the mechanisms bacteria use to communicate and compete with each other. Using model systems—specifically, species of soil bacteria, Bacillus and Streptomyces—and several experimental approaches, we aim to discover new interaction mechanisms, which in turn reveal the remarkable abilities bacteria have to sense and respond to other species.
Bacteria mobilizing bacteria

A two-species interaction between bacteria.: Streptomyces venezuelae–six spots in a horizontal line and Bacillus subtilis–six spots on a vertical line. When close to S. venezuelae, the B. subtilis becomes mobile, spreading to form an X pattern. See mSphere 3 (2018).
Using different species of Streptomyces, we observe that B. subtilis often mobilizes a population of cells to expand across a surface of agar. We selected one species and identified the inducer as the classic antibiotic, chloramphenicol, from S. venezuelae. Using this model interaction, we found that B. subtilis has an early warning system for the detection of competitors. We are investigating the mechanism that enables B. subtilis to detect chloramphenicol and other inducers. We are also investigating how B. subtilis switches from normal growth to expansion of the population, which is a mechanism of survival when confronted with antibiotic-producing competitors. Finally, we are using mobilization as a bioassay to detect and identify new inducer metabolites from Streptomyces spp. We will determine how the new inducers function, and if new antibiotics are among them.
Y. Liu, S. Kyle, P. D. Straight, Antibiotic stimulation of a bacillus subtilis migratory response. mSphere 3 (2018).
The secondary metabolome of Streptomyces
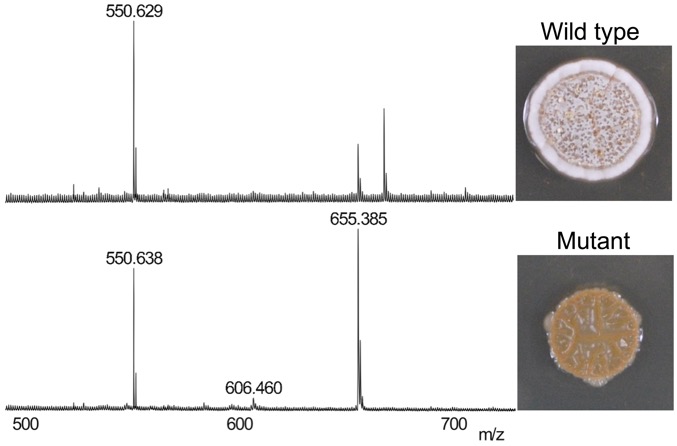
Disruption of secondary metabolite synthesis generates phenotypes, which are observed using MALDI-mass spectrometry (left) and colony morphology (right). We use this approach to discover new metabolites for bacterial growth and competitive fitness. (Unpublished data.)
Many species of bacteria produce bioactive metabolites that perform important functions in nature but are mysterious in laboratory cultures. Historically, the secondary metabolites of interest have been antibiotics. Thus, research in this field focuses primarily on antibiotic activities, leaving the majority of secondary metabolites with no known function. We are developing and using streamlined genetic approaches to disrupt the secondary metabolome of Streptomyces spp. in order to uncover new metabolites, their unknown functions, and the undiscovered antibiotics that may be hidden within secondary metabolome.
S. R. Barger, B. C. C. Hoefler, A. Cubillos-Ruiz, W. K. Russell, D. H. Russell, P. D. Straight, Imaging secondary metabolism of Streptomyces sp. Mg1 during cellular lysis and colony degradation of competing Bacillus subtilis. Anton. Leeuw. Int. J. G. 102, 435–445 (2012).
Biosynthetic Pipelines for Antibiotic Production by Bacteria
Biosynthetic pipelines for antibiotic production. A single species of bacteria may produce several different antibiotics. Their biosynthesis requires coordination of gene expression, enzyme assembly, central metabolism for precursors, and export machinery for delivery. We use combined approaches of light and electron microscopy with biochemical and genetic experiments to identify the elements of individual pipelines and their mechanisms of assembly in the bacterial cell.
One example of a biosynthetic pipeline is production of the antibiotic bacillaene by Bacillus subtilis. Bacillaene is synthesized by a megacomplex of enzymes. How these enzymes associate with the plasma membrane to export bacillaene is not known. We use genetic approaches to determine the cellular requirements for bacillaene export. A second example is the antibiotic family of linearmycins. We discovered that the linearmycins are packaged into extracellular vesicles by Streptomyces sp. Mg1. We are using this system to understand how the vesicles are produced, loaded with the linearmycins, and released across the cell envelope of these
Gram-positive bacteria. These studies will provide insights into the cellular constraints for antibiotic production that will facilitate bacterial engineering approaches to antibiotic discovery.
B. C. Hoefler, R. M. Stubbendieck, N. K. Josyula, S. M. Moisan, E. M. Schulze, P. D. Straight, A link between linearmycin biosynthesis and extracellular vesicle genesis connects specialized metabolism and bacterial membrane physiology. Cell Chem. Biol. 24, 1238–1249 (2017).
Metabolites and enzymes in competitive interactions
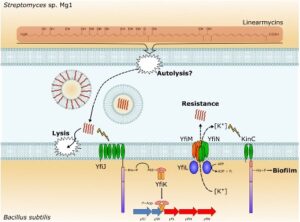
Linearmycins cause lysis of B. subtilis cells, but resistant cells emerge due to mutations that activate the LnrJK (YfiJK) two-component signaling system that controls an ABC transporter, LnrLMN (YfiLNM). LnrLMN provides linearmycin resistance and biofilm formation by an unknown mechanism.
See J. Bacteriol. 199 (2017).
Our studies using bacterial interactions are building a deeper understanding of bacterial competitive functions and fitness. Bacteria that produce bioactive metabolites may produce many of them at the same time. They may also produce and secrete enzymes that interact with metabolites of other organisms. These processes are integral to bacterial competition. Through in-depth studies of two species interactions, we are discovering layers of complexity in bacterial competition. Our data enables us to reconstruct elaborate views of the functions at work during competition, which we believe will better reflect the nature of bacterial interactions and community formation.
R. M. Stubbendieck and P. D. Straight, Escape from lethal bacterial competition through coupled activation of antibiotic resistance and a mobilized subpopulation. PLoS Genetics 11 (2015).
Publications
- View publications on PubMed