
-
Hays Rye
- Associate Professor, Biochemistry and Biophysics
- Focus Area: molecular chaperones, protein folding, protein quality control, disaggregation, membrane dynamics, membrane fission, nanoparticle dynamics, single molecule fluorescence
- Office:
- BICH 239A
- Email:
- [email protected]
- Phone:
- 979-862-1123 Lab phone: 979-862-1125
Education
- Undergraduate Education
- B.S. Rice University (1989)
- Graduate Education
- Ph.D. University of California, Berkeley (1995)
- Postdoc. Yale University (2000)
Areas of Expertise
- Protein folding and molecular chaperones
- Protein disaggregation and quality control
- Membrane fission
- Nanoparticle dynamics
- Fluorescence spectroscopy
- Rapid-mixing and enzyme kinetics
- Single-molecule and single-particle fluorescence
Professional Summary
The Rye lab is broadly interested in understanding how cells control the assembly and disassembly of important biological nanoparticles like protein aggregates and membrane transport vesicles. Specialized molecular machines powered by the energy stored in ATP or GTP are required for these activities and regulate many aspects of basic cellular biology. Our goal is to develop detailed mechanistic insights into these molecular machines and understand why their dysfunction leads to the wide range of diseases that spring from aberrant protein assembly and membrane dynamics.
Development of single particle analysis methods
The formation and disassembly of macromolecular particles are ubiquitous and essential features of all living organisms. Additionally, diseases are often associated with biologically active nanoparticles, such as the formation of toxic protein aggregates in diseases of protein misfolding and the growth of infectious viral particles. Simultaneously, the heterogeneous and dynamic nature of biologically active particles can make them exceedingly challenging to study. To tackle this problem, we are developing new techniques, e.g., burst analysis spectroscopy (BAS). We combine these methods with other single-molecule and single-particle approaches to uncover unique nanoparticle dynamics in cell biology.
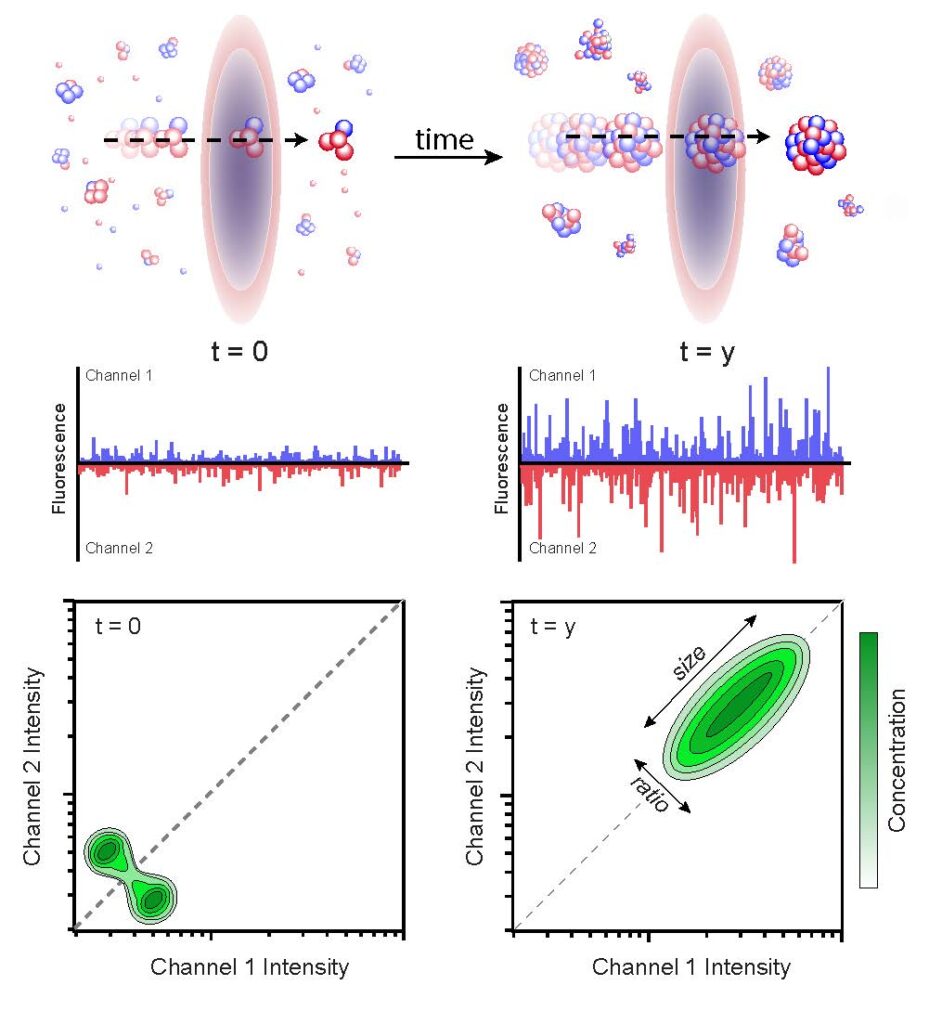
Burst analysis spectroscopy (BAS) measures the population-resolved kinetics of biological nanoparticle assembly and disassembly. Fluorescent particles are advectively flowed through one or more focused laser beams.
See PNAS 105, 14400–14405 (2008).
See: Puchalla, J., et al., PNAS 105, 14400–14405 (2008) and D. Shoup, et al., Biophys. J. 120, 2192-2204 (2021).
Protein disaggregation and molecular chaperones
The misfolding and aggregation of essential cellular proteins is a fundamental problem for all living organisms. Aggregation of even non-essential proteins can lead to debilitating diseases like type II diabetes, Alzheimer’s, Huntington’s, and Parkinson’s. Importantly, protein folding and aggregation are heavily influenced by the cellular protein quality control machinery involving networks of molecular chaperones. Precisely how different chaperone systems cooperate in dismantling and reactivating aggregated proteins and how molecular chaperone action affects disease progression, is not well understood. We use a variety of approaches, including novel single-particle assays like BAS, to study the disaggregate systems of bacteria and yeast. Our goal is to develop a mechanistic understanding of how different molecular chaperone networks recognize and dismantle protein aggregates.
All Publications
- View publications on Google Scholar