-
Mingyuan Zhu
- Assistant Professor
- Focus Area: Crop root development, Circumnutation, Environmental stress resilience of crops, single-cell transcriptomics
- Office:
- BICH 305A
- Email:
- [email protected]
- Phone:
- 979-314-8320
Education
- Undergraduate Education
- B.S. Life Sciences, Tsinghua University (2013)
- Graduate Education
- Ph.D. Plant Biology, Cornell University (2020)
- Postdoc. Rice root development, Duke University (2025)
Areas of Expertise
- Rice root growth patterns
- Single-cell RNA sequencing (scRNA-seq)
- Spatial transcriptomics
- Live imaging and quantitative cellular growth analysis
- Automatic crop seedling phenotyping
Professional Summary
In Zhu lab, we seek to comprehensively understand how organisms dynamically adapt their growth in response to environmental changes. We use crop roots as the model system.
One key environmental stress that developing roots encounter is physical obstacles blocking the root penetration. This is an urgent issue for global food security as mechanical soil stresses have greatly contributed to the degradation of over 30% of arable land, causing annual economic losses of $6.3 to $10.6 trillion.
Rice roots navigate through obstacles using one internal growth pattern known as circumnutation, defined as the helical movement of the root tip. This phenomenon, which is widespread across many plant species, has been mathematically described by two key parameters: frequency, the speed at which the root tip completes one circumnutation cycle, and amplitude, the maximum distance the root tip moves away from the central axis. Circumnutation frequency correlates with root penetration capacity while circumnutation amplitude enhances root navigation ability. Several key questions remain unanswered: Can plant roots adjust frequency and amplitude for better adaptation to environmental stresses? If so, what molecular regulators and cellular parameters govern these adjustments?
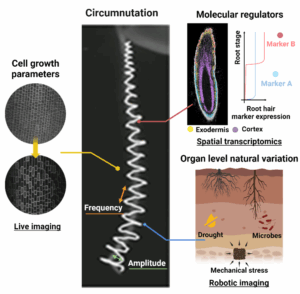
Overview of the research @ Zhu Lab
We are conducting multi-scale studies to identify the molecular regulators and cellular parameters that control circumnutation frequency and amplitude, and to assess organ-level natural variations in circumnutation.
DIRECTION1: The molecular regulators controlling circumnutation
We aim to identify molecular regulators of circumnutation frequency and amplitude through single-cell transcriptomics and CRISPR-based screening.
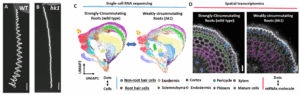
DIRECTION2: The molecular regulators controlling circumnutation
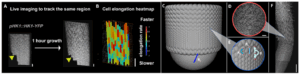
We aim to study cellular parameters regulating circumnutation frequency and amplitude by integrating quantitative live imaging (Panel A, B) with computational modeling (Panel C, E).
DIRECTION3: The organ-level environment driven variations in circumnutation
We aim to characterize organ-level circumnutation variations driven by environmental factors and also to further explore the agricultural significance of circumnutation with Robotic imaging.