
-
Xiuren Zhang
- Christine Richardson Endowed Professor of Agricultural Biology, EDGES Fellow, Presidential Impact Fellow, AgriLife Faculty Fellow, Professor, Biochemistry & Biophysics, Biology, Molecular & Environmental Plant Sciences Focus Area: RNA silencing, RNA structure and remodeling, ribonucleoprotein complexes, epi-transcriptome, epigenetics, host-virus interaction
- Focus Area: RNA silencing, RNA structure and remodeling, ribonuceloprotein complexes, epitranscriptome, epigenetics, host-virus interaction, geminivirus, cucumber mosaic virus
- Office:
- BICH 316A
- Email:
- [email protected]
- Phone:
- 979-314-8292
Education
- Undergraduate Education
- B.S. Hefei College of Economy and Technology, (1989)
- Graduate Education
- Ph.D. Cornell University, (2003)
- Postdoc. Rockefeller University, (2008)
Areas of Expertise
- Molecular Genetics
- RNA biology
- Posttranscriptional processing of RNA
- Posttranslational modification
- Host-microbe interaction
Professional Summary
Our research has the goal to identify new components of RNA silencing pathways and to study their functions and mechanisms in plants, as well as to decode the genetic information and biology hidden in the RSS and REM in selected model organisms. Additionally, we study host/virus interplay at an epigenetic level. We apply a combination of genetic, molecular, biochemical, and the state-of-art-mics approaches to study our projects.
RNA silencing

Genetic screening of new components in RNA silencing pathway.
See Cell 145, 242–256 (2011).
In the central dogma, RNA bridges the flow of genetic information from DNA to protein. Recent years have seen an explosion in the discovery of new species of RNAs and RNA-mediated processes. One group of tiny regulatory RNA molecules called miRNA or siRNA can guide RNA silencing to control various aspects of biology in eukaryotes including growth and development, stress responses, and antiviral defense. miRNA and sRNA are produced by Dicer or Dicer-like proteins, and loaded into Argonaute (AGO)-centered RNA-induced silencing complexes (RISCs). The RISCs execute the repressive or regulatory functions on target genes at transcriptional or post-transcriptional levels. One goal of our research is to identify new components of RNA silencing pathways and to study their functions and mechanisms in plants.
H. Zhu, F. Hu, R. Wang, X. Zhou, S.-H. Sze, L. Wen Liou, A. Barefoot, M. Dickman, X. Zhang, The Arabidopsis Argonaute 10 specifically recruits miR166/165 to maintain shoot apical meristem. Cell 145, 242–256 (2011).
RNA structural remodeling and epi-modifications
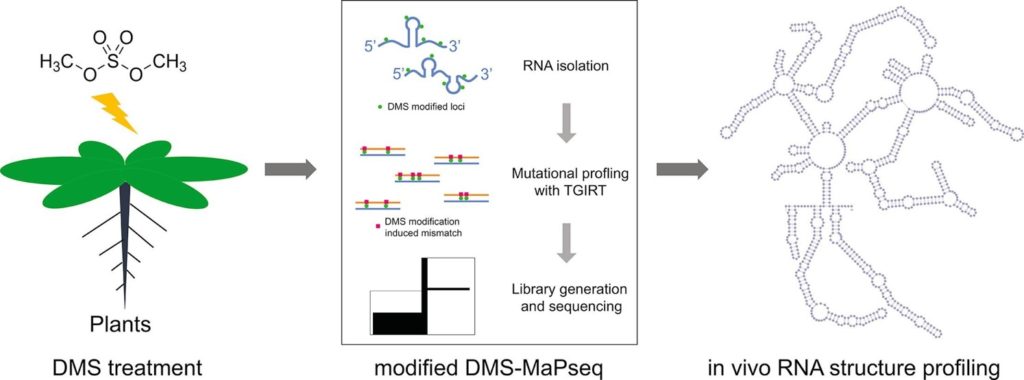
Genome-wide probing and functional analysis of RNA secondary structures.
See Methods 155, 30–40 (2019).
RNA is now also known to influence many aspects of biology through activities that are attributable to its secondary structure (RSS). In fact, RSS contains a new set of information code that is interpreted and processed by specialized protein complexes to regulate transcription, splicing, translation, localization, phase transition, and turnover. RSS can be further remodeled to meet specific functional needs in physiological conditions. Moreover, many RNA species undertake posttranscriptional RNA editing and/or epi-modification (REM) which even add more complexity to regulate gene expression and biological functions. Thus, another aspect of our research is to decode the genetic information and biology hidden in the RSS and REM in selected model organisms.
Z. Wang, Z. Ma, C. Castillo-González, D. Sun, Y. Li, B. Yu, B. Zhao, P. Li, X. Zhang, SWI/SNF subunit CHR2 remodels pri-miRNAs via SE to inhibit miRNA production. Nature 557, 516–521 (2018).
Host/virus interaction
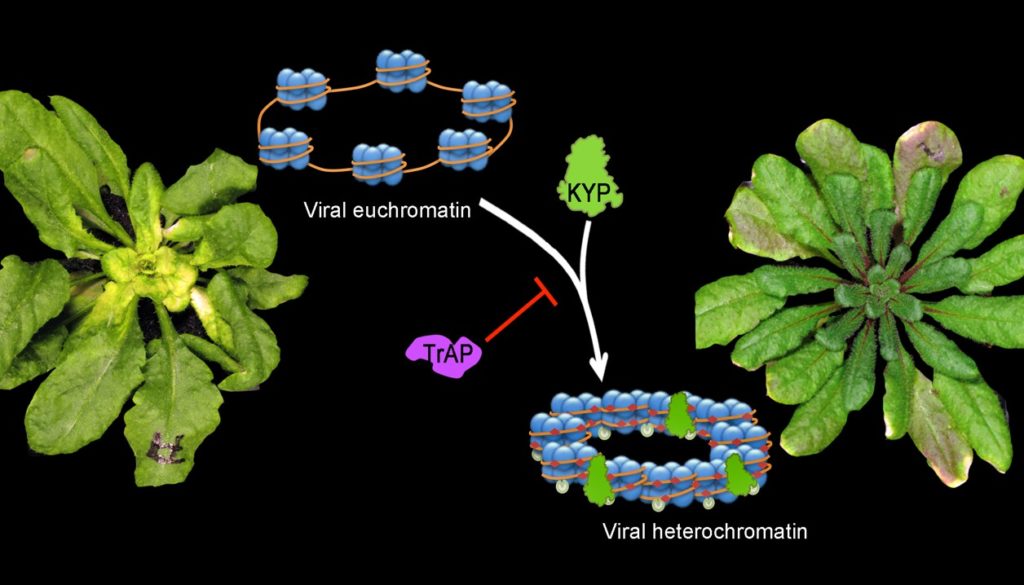
Epigenetic suppression by the viral protein TrAP.
See eLife 4, (2015).
Plants utilize RNA silencing to defend themselves from exogenous nucleic acid invaders (i.e., viruses). As an anti-host defense mechanism, viruses encode suppressors that can inhibit RNA silencing responses to evade host surveillance. Although our knowledge of host/virus interaction has dramatically expanded at the post-transcriptional gene silencing level, our understanding of epigenetic silencing and viral suppression is still very poor. We use the mode DNA virus Geminivirus among other viruses to study host/virus interplay at an epigenetic level.
C. Castillo-González, X. Liu, C. Huang, C. Zhao, Z. Ma, T. Hu, F. Sun, Y. Zhou, X. Zhou, X.-J. Wang, X. Zhang, Geminivirus-encoded TrAP suppressor inhibits the histone methyltransferase SUVH4/KYP to counter host defense. eLife 4, (2015).
Z. Wang Z, C. Castillo-González, C. Zhao, CY Tong, C Li, S Zhong Z, Liu, K Xie, J Zhu, Z Wu, X Peng, Y Jacob, SD Michaels, SE Jacobsen, X Zhang. H3.1K27me1 loss confers Arabidopsis resistance to Geminivirus by sequestering DNA repair proteins onto host genome. Nat Commun. 2023 Nov 18;14(1):7484.
Publications
- View publications on PubMed