
-
Junjie Zhang
- Professor, Biochemistry and Biophysics
- Focus Area: Translation in mycobacteria, intoxication and neutralization of the toxins from clostridium difficile, single-stranded RNA bacteriophages and their applications for RNA delivery
- Office:
- BCBP 410
- Email:
- [email protected]
- Phone:
- 979-314-8291
Professional Summary
The living cell contains a collection of molecular machines to grow and function. These machines include the ribosomes, the chaperons, the proteasomes and other enzymes. Malfunction of these machines are related to many human diseases. Elucidation of their three-dimensional (3D) structures is essential to understanding how these machines work in the cell and eventually to treat those related diseases. We use an experimental technique called cryo-electron microscopy (cryo-EM) to image these cellular machines in their native environment at liquid nitrogen temperatures.
Mycobacterial ribosomes
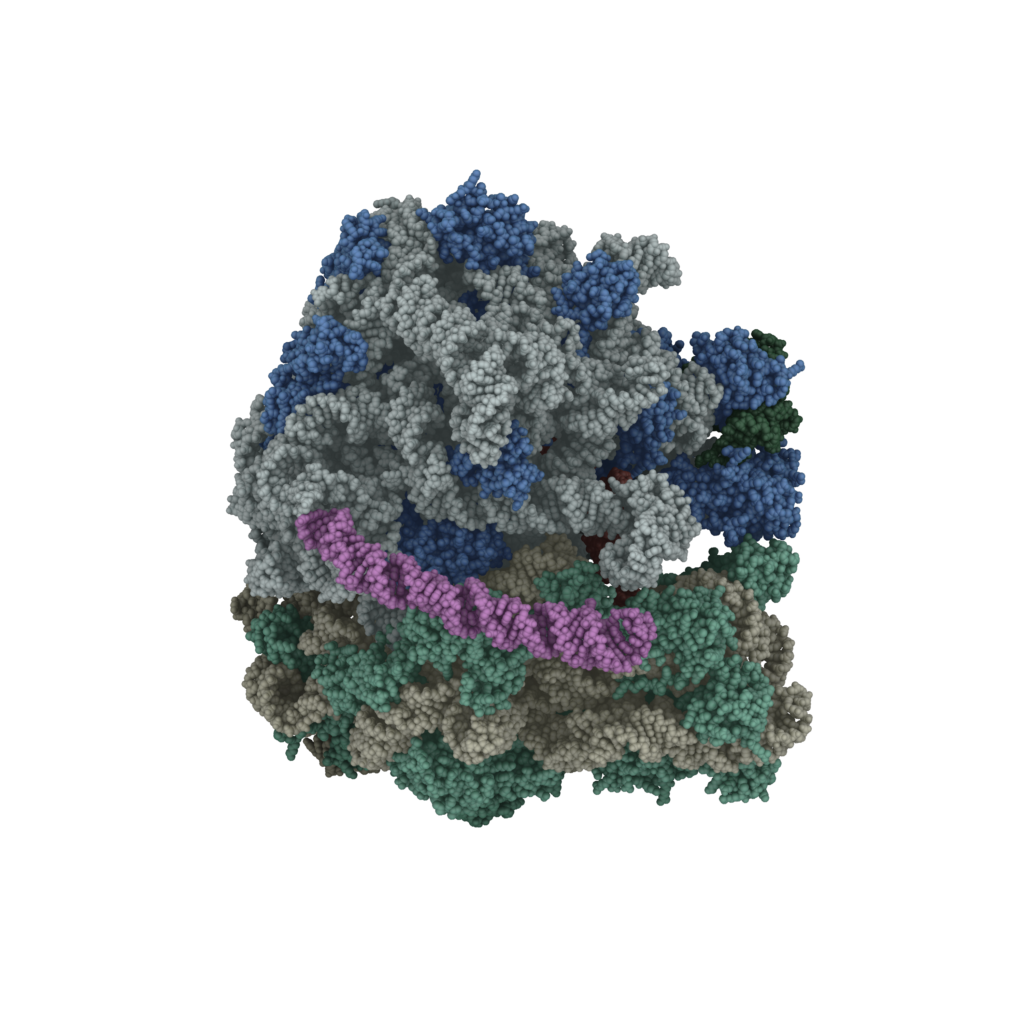
Cryo-EM structure of the Mycobacterium tuberculosis ribosome.
See Nucleic Acids Res. 45,10884–10894 (2017).
Tuberculosis (TB) is a chronic infectious disease caused by Mycobacterium tuberculosis (Mtb). In 2015, TB led to ~1.4 million deaths worldwide. The success of Mtb as a pathogen is largely attributable to its ability to persist in the host and to develop resistance to antibiotics. The Mtb ribosome is a major target for treating TB. The primary goals of this project are to study the mechanism of how the novel structural features of Mtb ribosomes impact protein synthesis and to provide structural information that facilitates the development of Mtb-specific therapeutics. This project is supported by the NIH grant P01AI095208 and the Welch foundation grant A1863.
K. Yang, J. Chang, Z. Cui, X. Li, R. Meng, L. Duan, J. Thongchol, J. Jakana, C. M. Huwe, J. C. Sacchettini, J. Zhang, Structural insights into species-specific features of the ribosome from the human pathogen Mycobacterium tuberculosis. Nucleic Acids Res. 45,10884–10894 (2017).
Single-stranded RNA bacteriophages

Cryo-EM structure of a single-stranded RNA bacteriophage.
See Proc. Natl. Acad. Sci. USA 113, 11519–11524 (2016).
The single-stranded ribonucleic acid (ssRNA) phages are small viruses which infect their host via retractile pili. This project focuses on the structural mechanisms by which ssRNA phages assemble themselves to recognize and destroy their host cells. This is important for a number of reasons: (a) understanding how ssRNA phages recognize and kill bacteria could lead to more specific and potent antibiotics; (b) understanding how ssRNA phages assemble could expand its application for packaging and delivery of other engineered RNA molecules with therapeutic and biotechnological interests; (c) many of the fundamental processes involved are mirrored in human RNA viruses and in important processes in human cell biology; and (d) More ssRNA phages are being discovered and investigated as potential antibacterial agents, so understanding the fundamentals of the ssRNA phage infection cycle is critical. This project is supported by the NSF grant 1902392.
R. Meng, M. Jiang, Z. Cui, J. Chang, K. Yang, J. Jakana, X. Yu, Z. Wang, B. Hu, J. Zhang, Structural basis for the adsorption of a single-stranded RNA bacteriophage. Nat. Commun. 10 (2019).
K. V. Gorzelnik, Z. Cui, C. A. Reed, J. Jakana, R. Young, J. Zhang, Asymmetric cryo-EM structure of the canonical Allolevivirus Qβ reveals a single maturation protein and the genomic ssRNA in situ. Proc. Natl. Acad. Sci. USA 113, 11519–11524 (2016).
Clostridium difficile toxins
C. difficile infection (CDI) is the leading cause of hospital-acquired infectious diarrhea and colitis, claiming the lives of ~30,000 Americans each year. The primary cause of symptoms from CDI is due to its secreted toxins, which affect the host cells in the colon. The goal of the current project is to establish a structural framework to reveal how the C. difficile toxins intoxicate the host cell and can be neutralized by engineered non-antibody protein therapeutics, which will provide a novel approach to treat CDI diseases. This project is supported by the NIH grant R21AI137696.
R. Simeon, M. Jiang, A. M. Chamoun-Emanuelli, H. Yu, Y. Zhang, R. Meng, Z. Peng, J. Jakana, J. Zhang, H. Feng, Z. Chen, Selection and characterization of ultrahigh potency designed ankyrin repeat protein inhibitors of C. difficile toxin B. PLoS Biol. 17 (2019)