
-
Lanying Zeng
- Professor, Biochemistry and Biophysics
- Focus Area: Cellular decision making, cellular dynamics, quantitative biology, biophysics, bacteriophage, bacterial pili, high-resolution and super-resolution fluorescence microscopy, mathematical modeling
- Office:
- BICH 419A
- Email:
- [email protected]
- Phone:
- 979-845-2961
- Graduate Education
- Ph.D. in Theoretical and Applied Mechanics, University of Illinois at Urbana-Champaign, Urbana, IL (2007)
- Postdoc. in Quantitative Biology and Biophysics, University of Illinois at Urbana-Champaign, Urbana, IL (2007-2011)
Areas of Expertise
- Bacteriophage
- Quantitative biology
- Biophysics
- Fluorescence microscopy
- Mathematical modeling
Professional Summary
We are interested in understanding various dynamical processes in the infection cycles of phages, viruses of bacteria. The studies are aimed at uncovering fundamental mechanisms in these processes at the single-cell, single-molecule level, which can shed important light on antibacterial prophylaxis and therapy. The major tools used for our studies are high-resolution and super-resolution fluorescence microscopy combined with mathematical modeling.
Cell-fate decision making
A 4-color system reports different cell fates upon infection of phage lambda. See Nat. Commun. 8 (2017).
The system of E. coli and its virus, phage lambda, has long served as a paradigm for cell-fate bifurcation. As a temperate phage, upon infecting an E. coli cell, lambda can choose either the lytic (virulent) pathway producing ~100 new progeny particles and lysing the cell, or the lysogenic (dormant) pathway with its DNA integrated into the host chromosome and replicating along with the host. Recent single-cell, single-molecule studies have yielded many interesting findings including individual phage voting, interplay of phage competition and cooperation. We are interested in formulating a quantitative picture of lysis-lysogeny decision making of lambda. We have also expanded our study to another model system, phage P1, well known for its high rate of generalized transduction. P1 has unusual properties of lysis-lysogeny. We aim to elucidate the fundamental mechanisms of these model systems, which will shed important light on phage therapy treating multi-drug resistant bacteria and how other viruses operate in the host including higher organisms.
Zhang, K., Pankratz, K., Duong, H., Theodore, M., Guan, J, Jiang, A., Lin, Y. and Zeng, L., Interactions Between Viral Regulatory Proteins Ensure an MOI-Independent Probability of Lysogeny during Infection by Bacteriophage P1, mBio 12(5): e01013-21 (2021)
Trinh, J.T., Shao, Q., Guan J., and Zeng, L., Emerging Heterogeneous Compartments by Viruses in Single Bacterial Cells, Nature Communications 11, 3813 (2020)
Shao, J. T. Trinh, L. Zeng, High-resolution studies of lysis-lysogeny decision making in bacteriophage lambda. J. Biol. Chem. 294, 3343–3349 (2019).
Trinh, J.T., Székely, T., Shao, Q., Balázsi, G., and Zeng, L., Cell Fate Decisions Emerge as Phages Cooperate or Compete inside their Host, Nature Communications 8, 14341 (2017)
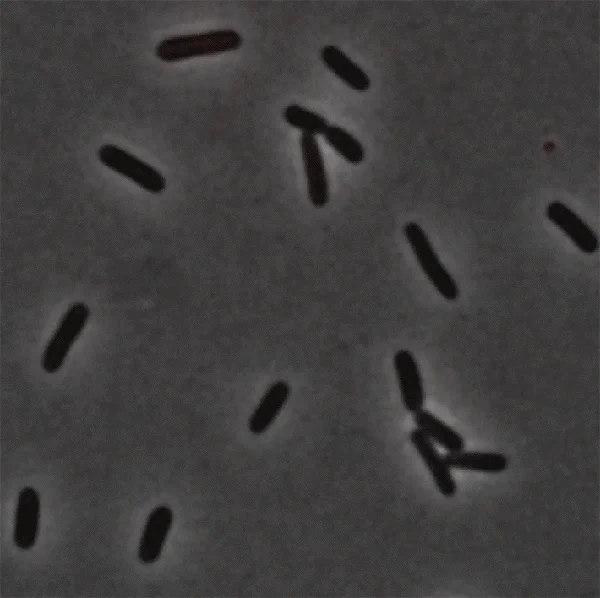
Infection cycle of ssRNA phages

The E. coli F-pili are detached due to the infection of ssRNA phage. See PNAS 117(41), 25751-25758 (2020).
ssRNA phages are small viruses which infect bacteria through retractile pili. We are particularly interested in the entry dynamics of genomic RNA from the phage capsid to the cell, virus replication and assembly inside the cell, and the impact on the retractile pili as a result of ssRNA phage infection. We aim to understand these fundamental processes and to engineer these phages as novel antibacterials.
Thongchol, J.#, Yu, Z.#, Harb, L., Lin, Y., Koch, M., Theodore, M., Narsaria, U., Shaevitz, J., Gitai, Z., Wu, Y., Zhang, J.*, and Zeng, L.*, “Removal of Pseudomonas Type IV Pili by a Small RNA Virus”, Science 384, eadl0635 (2024) #, co-first author. *, co-corresponding author.
Harb, L., Chamakura, K., Khara, P., Christie, P.J., Young, R., and Zeng, L., ssRNA Phage Penetration Triggers Detachment of the F-pilus, PNAS 117(41), 25751-25758 (2020)
All Publications
- View publications on PubMed