
-
Ry Young
- University Distinguished Professor; Professor Emeritus, Biochemistry & Biophysics, Biology
- Focus Area: bacteriophage and bacterial molecular biology, membrane proteins, prokaryotic and phage bioinformatics and genomics, genetic engineering, phage therapeutics (NOT CURRENTLY TAKING GRADUATE STUDENTS)
- Office:
- BICH 443A
- Email:
- [email protected]
- Phone:
- 979-314-8299
Professional Summary
As a founding lab of the Center for Phage Technology, our group is committed to phage and bacterial molecular genetics and bioinformatics. Our central focus is on the mechanisms by which phage cause lysis of their bacterial hosts. We bring a wide range of techniques to bear on this and related problems, including molecular genetics, cell biology, biochemistry and genomics, especially as leveraged by our collaborators in the CPT. Besides contributing to a mechanistic understanding of viral-host interactions, our studies often reveal new fundamental properties of proteins and of the pathways and mechanisms that underlie the homeostasis of the bacterial envelope. We are also interested in the applications of phage therapeutics to the fight against the current crisis of multi-drug resistant bacterial infection.
Single gene lysis
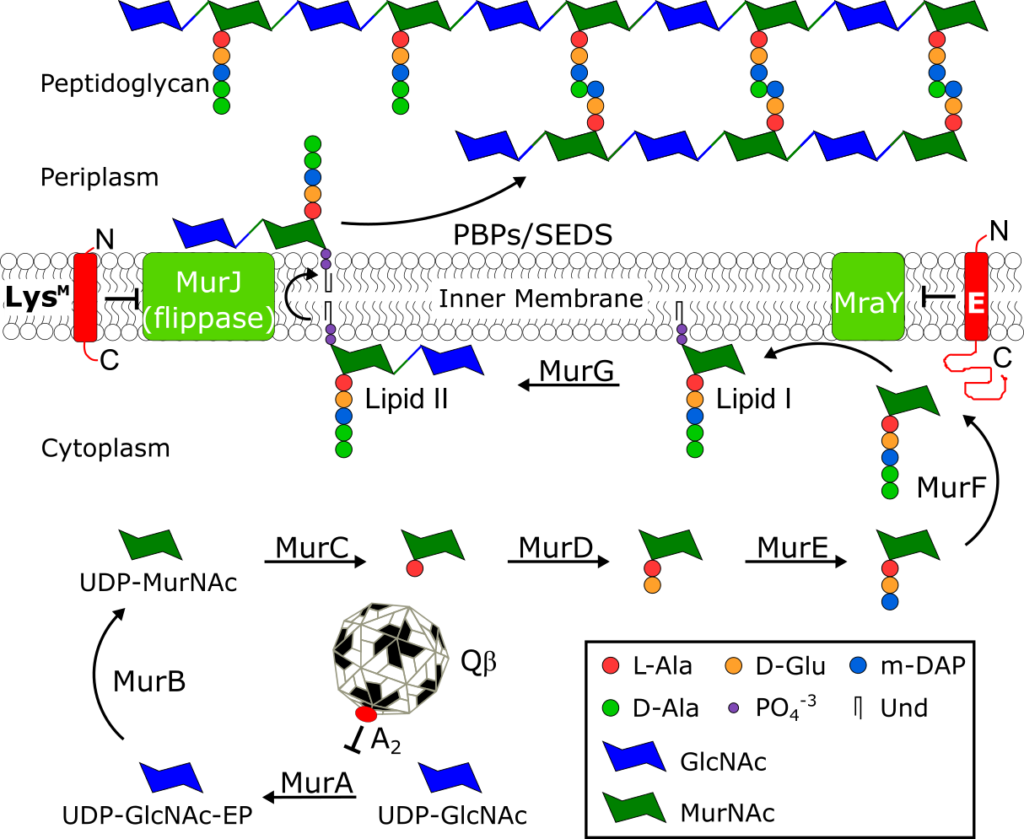
The peptidoglycan biosynthesis pathway and Sgl inhibitors.
See J. Biol. Chem. 294, 3350–3358.
Small lytic phages, microviruses (ssDNA) or leviviruses (ssRNA) encode a single gene to effect host lysis and the gene is designated as sgl (single gene lysis). The most studied sgls are gene E of the canonical microvirus φX174, lysM from ssRNA phage M, and genes A2 and L of the paradigm ssRNA phages Qβ and MS2, respectively. All four lysis proteins lack muralytic activity, and three, A2, E and LysM, have been shown to function as “protein antibiotics” by acting as inhibitors of conserved peptidoglycan (PG) biosynthesis enzymes, MurA, MraY, and MurJ. Recent global metagenomic efforts have uncovered thousands of new phage genomes, each with a potentially novel sgl and a novel mode of action. Studying these new sgl proteins could potentially lead to identification of new antibiotic targets.
K. R. Chamakura, R. Young, Phage single-gene lysis: finding the weak spot in the bacterial cell wall. J. Biol. Chem. 294, 3350–3358 (2019).
K. R. Chamakura, L.-T. Sham, M. R. Davis, L. Min, H. Cho, R. Ruiz, T. G. Bernhardt, R. Young, A viral protein antibiotic inhibits lipid II flippase activity. Nat. Microbiol. 2, 1480–1484 (2017).
Multi-Gene Lysis
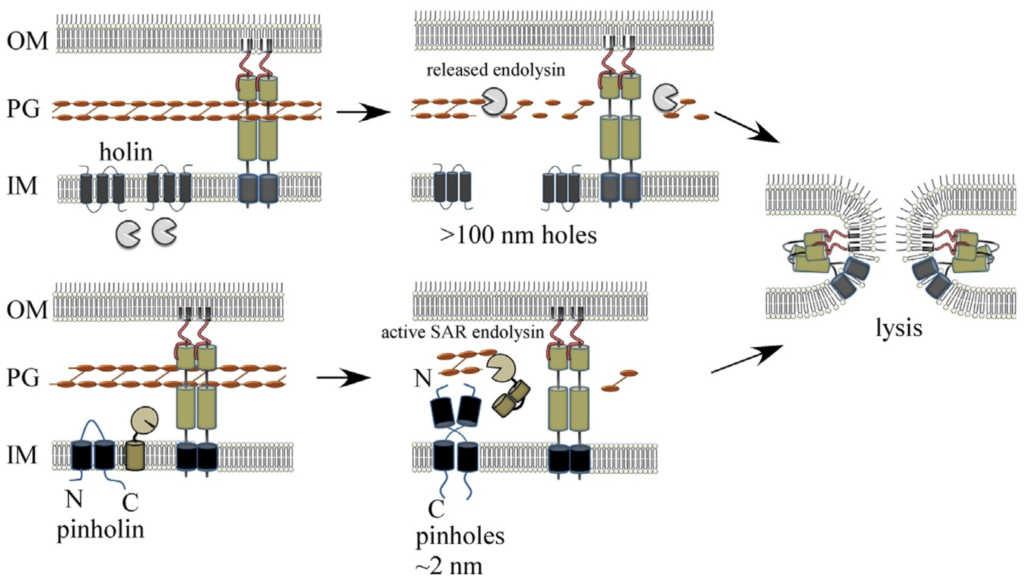
Model for spanin-mediated canonical holin-endolysin and pinholin-SAR endolysin phage MGL pathways.
See Adv. Virus Res. 103, 33–70 (2019).
Tailed phages use carefully timed multi-gene lysis (MGL) systems with proteins individually targeting the cellular barriers to assembled particle exit. Specifically, holins form holes in the inner membrane and cause instant cell death. The holes that collapse membrane potential can be small, such as the phage 21 pinholin, or micron-scale, as in the canonical phage lambda holin, which is large enough to allow proteins to pass into the periplasmic compartment. Phage-encoded peptidoglycan hydrolases called endolysins then degrade the cell wall. In Gram-negative hosts, phages also disrupt the outer membrane, usually with proteins called spanins, breaching the last barrier to virion release. We recently identified new phage lysis protein classes: disruptins are cationic peptides similar to mammalian antimicrobial peptides that breach the outer membrane, and releasins license dynamic membrane topology of endolysins. These studies expand our understanding of the diverse mechanisms used in MGL, the most common cytolytic event on earth.
K. To and R. Young, Probing the Structure of the S105 Hole. J. Bacteriol. 196, 3683–3689 (2014).
R. Kongari, J. Snowden, J. Berry, R. Young, Localization and regulation of the T1 unimolecular spanin. J. Virol. 92 (2018).