
-
Ann Wehman
- Assistant Professor
- Focus Area: Developmental genetics, Molecular cell biology, Membrane Trafficking, Extracellular Vesicles, Phagocytosis, Intracellular Clearance
- Office:
- BICH 332
- Email:
- [email protected]
- Phone:
- 979-314-8274
Education
- Undergraduate Education
- B.S. Biology, Massachusetts Institute of Technology, MA (1999)
- Graduate Education
- Ph.D. Genetics, University of California, San Francisco (2006)
- Postdoc. Developmental Genetics, New York University Medical Center (2006-2012)
Areas of Expertise
- Developmental genetics
- Molecular cell biology
- C. elegans
- Membrane Trafficking
- Extracellular Vesicles
- Phagocytosis
- Intracellular Clearance
Professional Summary
Membrane Dynamics
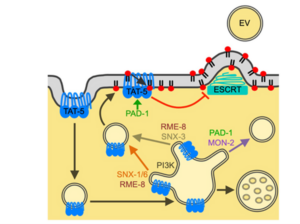
Katharina Beer et al., PNAS 2018
Extracellular Vesicles (EV)
We are using the power of C. elegans genetics to identify additional proteins that regulate EV budding. We revealed that conserved regulators of viral budding also have a role in EV budding in C. elegans, including the membrane-sculpting complex known as ESCRT. Our studies are building a pathway of proteins that regulate TAT-5 localization and activity and thereby modulate EV release, which are likely to be co-opted by viruses. The proteins we identify may be used to alter EV production in other systems, which could impact the availability of non-invasive biomarkers and have the potential to influence viral spread or disease state.
Studying the mechanisms of EV production has provided us with techniques to induce or prevent their formation. This allows us to test which signaling pathways require EVs for signaling to occur, as well as define other functional roles for EVs. We are studying how changing EV production or uptake affects conserved developmental and immune signaling pathways in C. elegans. Thus, our research aims to define the diverse functional roles of EVs, which are likely to be similar in humans.
Clearance by Phagocytosis
Tissues contain cell fragments and dying cells in addition to healthy cells. For example, cells release a remnant of the intercellular bridge after cell division as a 1 µm EV. Cells need to clear these debris from their environment by phagocytosis. Furthermore, the immune system needs to degrade cell debris in phagolysosomes to avoid generating an autoimmune response. We use elegans to study the mechanisms of EV and cell corpse uptake by phagocytosis and the steps of phagolysosomal clearance.
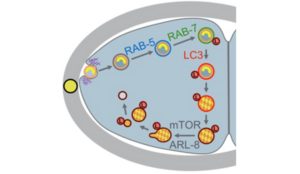
Gholamreza Fazeli et al., Cell Reports 2018
C. elegans embryonic cells take up mitotic midbody remnants released during cell division in addition to dying cells such as the meiotic polar body. These phagosomes mature similar to mammalian phagosomes and gradually acidify and degrade their cargo. Thus, we can use C. elegans to study the conserved mechanisms of EV signaling, the pathways regulating EV uptake, and use time-lapse imaging to determine the ultimate fate of engulfed cargos in a developing animal.
Lipid asymmetry also regulates this dynamic process and we are interested in the signaling roles of lipids as well as proteins and metabolites. Analyzing defects in EV uptake complements our studies on EV budding and will allow us to elucidate the interplay of lipids and lipid regulators during dynamic remodeling of the membrane. Studying the fate of EVs also provides important insights into the functional roles of EV, which are likely to be conserved in humans. Furthermore, understanding these mechanisms will help to identify key modulators of the immune response, which can be disrupted during pathogen infection or autoimmune disease.