
-
Josh Wand
- University Distinguished Professor and Head, Biochemistry & Biophysics, Professor, Biochemistry & Biophysics, Chemistry, Molecular & Cellular Medicine
- Focus Area: molecular recognition, drug discovery, protein dynamics, NMR spectroscopy, thermodynamics, protein engineering, protein biologics, ubiquitin E3 ligases, parkin, membrane proteins, prostate cancer, Parkinson’s disease (NOT CURRENTLY TAKING GRADUATE STUDENTS)
- Office:
- NMR N315A
- Phone:
- 979-314-8284 Research Office: (979) 314-8303
Education
- Undergraduate Education
- B.Sc. Hons. Biochemistry (1979)
- Graduate Education
- M.Sc. Chemistry (1981)
- Ph.D. Biophysics (1984)
- Postdoc. Biophysics (1985)
Areas of Expertise
- Molecular recognition
- Drug discovery
- Protein dynamics
- NMR spectroscopy
- Thermodynamics
- Protein engineering
- Protein biologics
- Ubiquitin E3 ligases
- Parkin
- Membrane proteins
- Prostate cancer
- Parkinson’s disease
Professional Summary
We are broadly interested in how the biophysical properties of proteins are manifested in their biological function. We are particularly engaged in trying to reveal the nature of internal protein motion and how this influences functions ranging from molecular recognition to allostery and catalysis. These basic ideas are being employed in a range of studies including protein engineering to optimize protein drugs, reverse micelle encapsulation to aid fragment-based drug discovery, understanding the regulation of Parkin, which is involved in mitophagy and early onset Parkinson’s Disease, and the enzyme AKR1C3, which is central to resistant forms of prostate cancer.
Molecular recognition by proteins
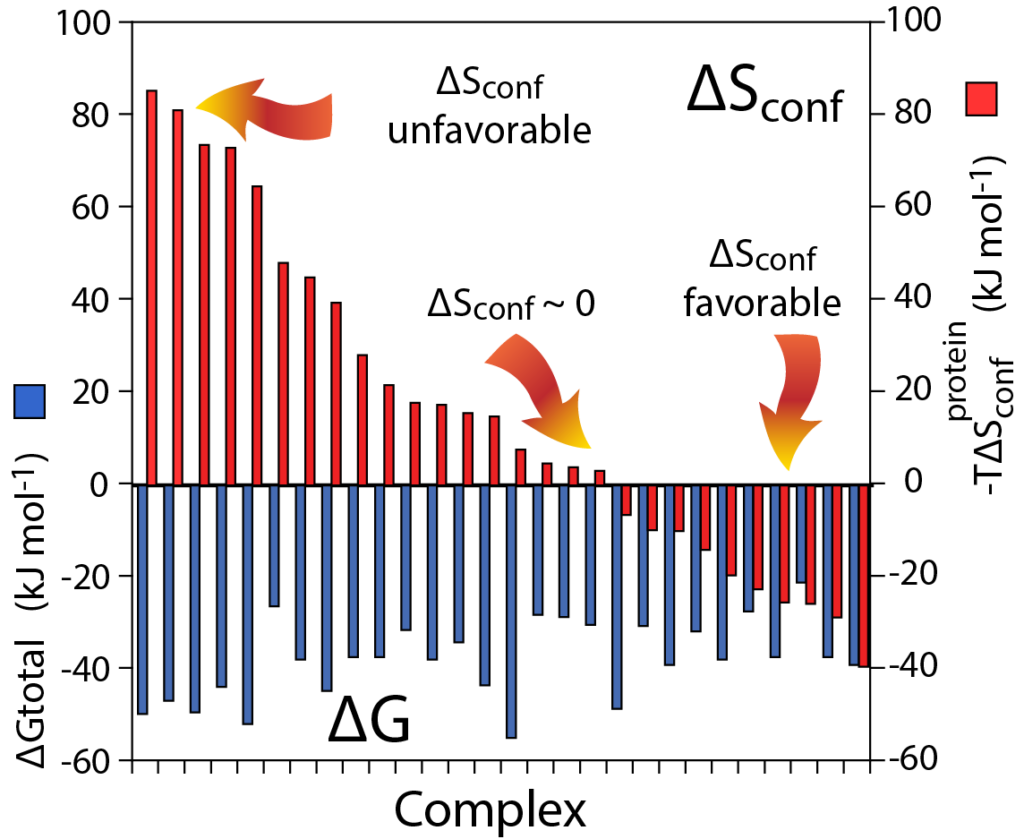
Proteins are internally dynamic – they change their structure all the time. This corresponds to conformational entropy (ΔSconf). We can now measure it by NMR and have found that ΔSconf often dominants molecular recognition by proteins, a fundamental element of molecular biology.
See PNAS 114, 4278–4280 (2017).
The popular view of protein thermodynamics and function has been largely driven by the exquisite detail of static structures, which is, unfortunately, a very enthalpic (energetic) view. This is only half the story since it is the free energy that is important. We are interested in the role of entropy is protein function. Historically it has been virtually impossible to measure the conformational entropy of proteins. We have developed experimental and analytical strategies to characterize internal motion of proteins and use this information as a quantitative proxy for conformational entropy. This breakthrough illuminated a whole new view of how proteins really work. For example, we have found a large and variable role for conformational entropy in the binding of ligands by proteins. Such a role for conformational entropy in molecular recognition has significant implications for enzymology, signal transduction, allosteric regulation and the development of protein-directed pharmaceuticals.

We have discovered that membrane proteins have extraordinary side-chain motion on the ps-ns timescale. The extensive side-chain motion about a highly rigid backbone scaffold is consistent with high conformational entropy and helps explains the stability of the folded state in the absence of the hydrophobic effect that dominates the stability of soluble proteins. See Angew. Chemie Int’l Ed. 59, (2020).
Applications of these fundamental concepts include using dynamics and entropy to optimize the affinity of protein biologics (protein drugs) for their targets. The emergence of protein biologics, particularly in the context of cancer immunotherapy, provides an important context for using dynamically-guided entropic manipulation of ligand binding affinity. Similarly, these ideas may help explain how complex enzymes, such as the ubiquitin E3 ligase Parkin, are allosterically regulated. We have recently discovered that integral membrane proteins display unique patterns of internal motion, which suggests that their functions are heavily influenced by changes in their conformational entropy. It turns out that integral membrane proteins are much more dynamic (entropic) than soluble proteins. This helps explain why they are so stable in the membrane without the benefit of the classic “hydrophobic effect.”
J. A. Caro, K. W. Harpole, V. Kasinath, J. Lim, J. Granja, K. G. Valentine, K. A. Sharp, A. J. Wand, Entropy in molecular recognition by proteins. Proc. Natl. Acad. Sci. USA 114, 6563–6568 (2017).
A. J. Wand and K. A. Sharp, Measuring entropy in molecular recognition by proteins. Annu. Rev. Biophys. 47, 41–61 (2018).
Protein hydration
Despite its importance, the interaction of between water and protein molecules remains a mystery. Over the past decade we have developed a strategy for the resurrection of a simple but flawed approach to characterizing protein hydration using solution NMR methods. It turns out that encapsulation of a protein within the water core of a reverse micelle eliminates the artifacts that have hindered the use of this NMR-based approach. With these barriers overcome we have found that protein hydration dynamics is remarkably variable across the surface of the protein. This feature has significant implications on the basic biophysical properties of proteins in solution and may have important impact on understanding the interaction of proteins with ligands including small molecule drugs.
N. V. Nucci, M. S. Pometun, A. J. Wand, Site-resolved measurement of water-protein interactions by solution NMR. Nat. Struct. Mol. Biol. 18, 245–249 (2011).
C. Jorge, B. S. Marques, K. G. Valentine, A. J. Wand, Characterizing protein hydration dynamics using solution NMR spectroscopy. Methods Enzymol. 615, 77–101 (2019).
Enhancing fragment-based drug discovery
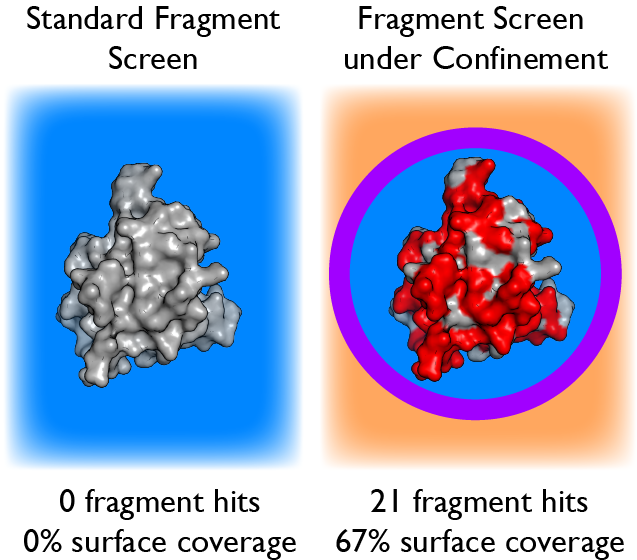
Encapsulation of protein molecules within the water core of a reverse micelle permits detection of extremely weak protein-ligand interactions by solution NMR spectroscopy. This enables a major advance in fragment-based drug discovery. Shown is the unprecedented surface coverage of interleukin-1β, which has been considered undruggable.
See ACS Chem. Biol. 14, 2224–2232 (2019).
We are using the confined space of the reverse micelle to facilitate NMR-based assays of so-called fragment libraries. Despite tremendous technological advances in drug discovery, rational de novo development of small molecule drugs is still difficult. High-throughput screening (HTS) of large natural product libraries thus remains the bedrock approach. HTS is unsatisfactory in many ways. Fragment-based drug discovery (FBDD) offers greater potential for the elaboration of hits to leads but has not penetrated the discovery space to the extent anticipated. This is because “fragments” are weak binders (Kd > 1 mM), which makes activity-based and standard biophysical assays unreliable, and the enormous potential of (FBDD) is lost. Our new reverse micelle encapsulation strategy is able to reliably detect very weak binding in a structural context. This capability could revolutionize early phase drug discovery. We are gearing exploring the potential of this approach to target enzymes that are involved in prostate cancer, COVID-19, breast cancer, and other human disease states.
B. Fuglestad, N. E. Kerstetter, S. Bédard, A. J. Wand, Extending the detection limit in fragment screening of proteins using reverse micelle encapsulation. ACS Chem. Biol. 14, 2224–2232 (2019).
Protein NMR methods
Our work is heavily based on the unique and spectacular capabilities of solution NMR spectroscopy. Along the way, we have developed the hardware used for high-pressure NMR; created novel NMR data processing strategies; developed new isotopic labeling methods; advanced applications of NMR in protein hydrogen exchange strategies; invented the reverse micelle encapsulation strategy for solution NMR; and so on. Current emphasis in on extending the capabilities of non-uniform data acquisition.
B. Fuglestad, B. S. Marques, C. Jorge, N. E. Kerstetter, K. G. Valentine, A. J. Wand, Reverse micelle encapsulation of proteins for NMR spectroscopy. Methods Enzymol. 615, 43–75 (2019).
J. A. Caro and A. J. Wand, Practical aspects of high-pressure NMR spectroscopy and its applications in protein biophysics and structural biology. Methods 148, 67–80 (2018).
A.C. Bishop, G. Torres-Montalvo, S. Kotaru, K. Mimun, A.J., Wand Robust automated backbone triple resonance NMR assignments of proteins using Bayesian-based simulated annealing. Nature Communications 14, 1556 (2023).
All Publications
- View publications on Google Scholar