
-
Cecilia Tommos
- Professor, Biochemistry and Biophysics
- Focus Area: protein electron transfer, proton-coupled electron transfer, PCET, amino-acid radicals, protein design, protein film voltammetry, unnatural (noncanonical) amino acids, chemical biology
- Office:
- NMR N315B
- Email:
- [email protected]
- Phone:
- 979-314-8296
Education
- Undergraduate Education
- B.S. (equivalent) Stockholm University, 1992
- Graduate Education
- Ph.D. Stockholm University, 1997
- Postdoc. University of Pennsylvania, 2000
Areas of Expertise
- Protein electron transfer (ET) and proton-coupled electron transfer (PCET)
- Protein radical chemistry
- Protein design and engineering, including using noncanonical amino acids
- Protein structural characterization using CD and NMR spectroscopic methods
- Protein film voltammetry
Professional Summary
Oxidoreductases use metallocofactors, organic molecules, and four amino acids (Tyr, Trp, Cys, and Gly) to perform electron transfer (ET) and proton-coupled electron transfer (PCET) reactions. These four amino acids serve as one-electron (radical) redox mediators in biocatalytic and multistep electron/hole transfer processes, some of which are essential to life on earth. There is also a sinister side to amino-acid ET/PCET since these reactions can be induced at oxidative stress conditions and cause significant cellular damage. It is extremely challenging to experimentally resolve the thermodynamic and kinetic redox properties of a single amino-acid residue. Consequently, key information to support the understanding of one of nature’s essential redox tools is missing. Our research focuses on developing a family of well-structured model proteins with the unique capability of providing detailed mechanistic information on Tyr (Y) and Trp (W) oxidation-reduction reactions.
The α3X model protein system
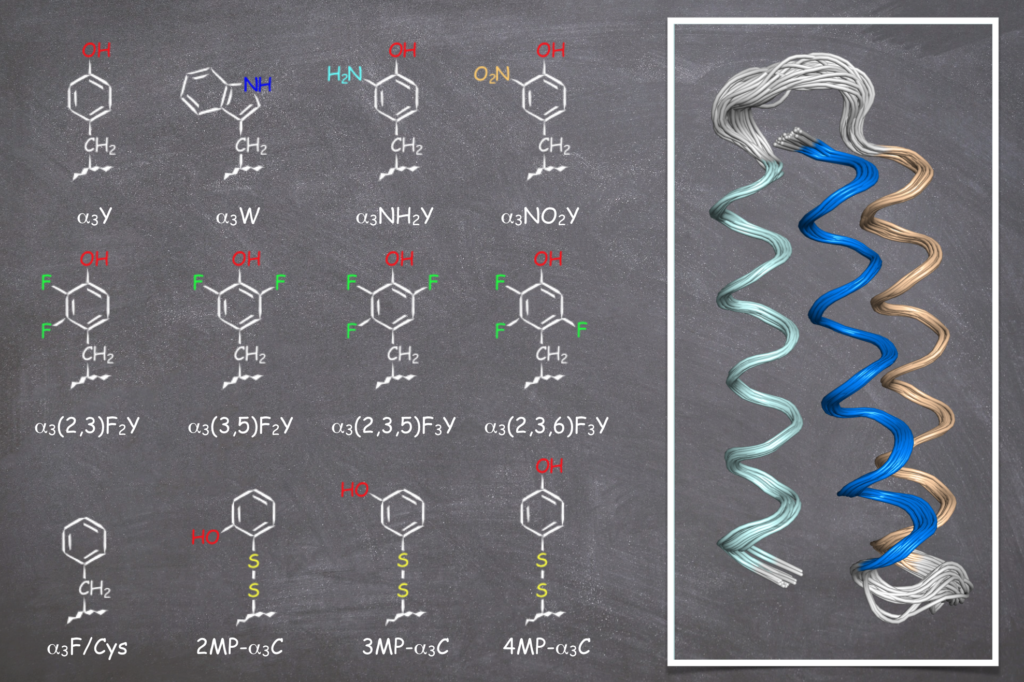
The present state of the a3X family of model proteins designed to study Y and W redox chemistry. The a3X proteins are based on a three-helix bundle (a3) with a single redox-active residue at interior position 32 (X32). Site 32 is occupied by Y32 (in a3Y), W32 (a3W), NH2Y32 (a3NH2Y), and so forth for the residues displayed in the figure above. See Tommos (2022) Annual Review of Biophysics, 51, 453–471.
We have made a family of well-structured model proteins specifically designed to study Y and W radical formation, transfer, and decay. This model system provides us with the rare opportunity to study amino acid radical reactions under fully reversible conditions inside a structurally well-determined protein environment. The α3X proteins were developed by combining protein design with site-specific incorporation of unnatural amino acids and detailed structural studies. We now aim to expand the α3X system by constructing well-structured proteins containing two redox-active residues (α3YX and α3WX, X = Y, W & Y/W analogs). These proteins will be explored to examine fundamental ET/PCET properties associated with Y/W-based radical transfer.
Tools to study Y/W oxidation-reduction
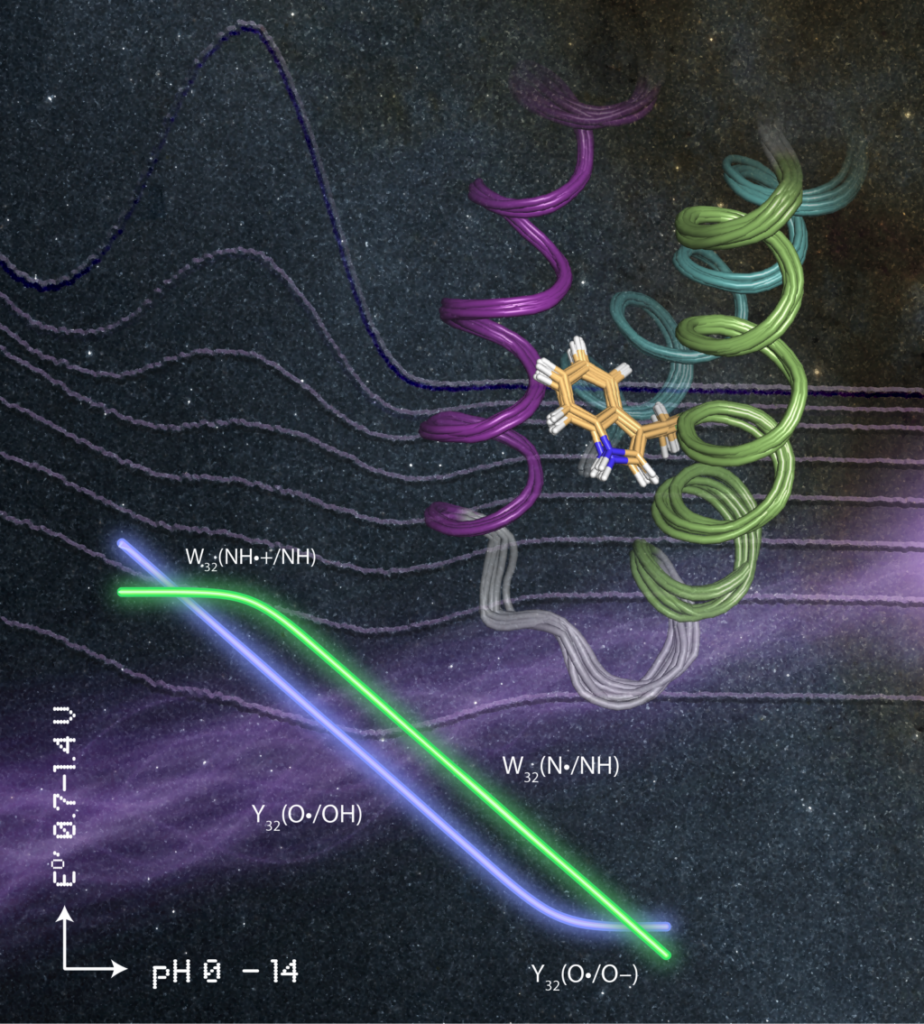
Artistic illustration of basic structural (solution NMR structure of α3W shown as a ribbon diagram), thermodynamic (E°’ vs. pH plots for Y32 and W32 shown in neon) and kinetic (transient absorption spectra following the decay of W32•) characterization of the α3X proteins.
See JACS 140, 185–192 (2018).
Mechanistic studies of the α3X proteins (and the α3XX proteins under construction) require a range of biophysically oriented approaches. These include obtaining essential thermodynamic information using a (very) high potential protein film voltammetry approach developed by the Tommos lab. In collaborative work, we use laser spectroscopy and theoretical methods to determine X32 radical formation and decay mechanisms. Through collaborative efforts, we also use advanced nuclear magnetic resonance (NMR) and electron paramagnetic resonance (EPR) spectroscopic methods to connect the redox properties of X32 with the structural and dynamical properties of the α3 protein scaffold. Additionally, we will label our proteins with ruthenium (Ru) complexes to study the PCET kinetics of light-triggered radical formation (in Ru-α3Y and Ru-α3W) and radical transfer (in Ru-α3YX and Ru-α3WX). The overall goal is to obtain a comprehensive and predictive understanding of Y and W radicals in biological ET/PCET processes.
B. W. Berry, M. C. Martínez-Rivera, C. Tommos (2012) Reversible voltammograms and a pourbaix diagram for a protein tyrosine radical. Proc. Natl. Acad. Sci. U.S.A. 109, 9739–9743.
K. R. Ravichandran, A. B. Zong, A. T. Taguchi, D. G. Nocera, J. Stubbe, C. Tommos (2017) Formal reduction potentials of difluorotyrosine and trifluorotyrosine protein residues: Defining the thermodynamics of multistep radical transfer. J. Am. Chem. Soc. 139, 2994–3004.
A. Nilsen-Moe, C. R. Reinhardt, S. D. Glover, L. Liang, S. Hammes-Schiffer, L. Hammarström, C. Tommos (2020) Proton-coupled electron transfer from tyrosine in the interior of a de novo protein: Mechanisms and primary proton acceptor. J. Am. Chem. Soc. 142, 11550–11559.
Tommos, C. (2022) Experimental characterization of aromatic amino acid free radicals in proteins, Annual Review of Biophysics, 51, 453–471.
All Publications
- View publications on PubMed