
-
Michael Polymenis
- Professor and Associate Head for Graduate Programs
- Focus Area: cell cycle, protein synthesis, ribosomal proteins, metabolism, cell size, longevity, yeast
- Office:
- BICH 333A
- Email:
- [email protected]
- Phone:
- 979-314-8287 Lab phone: 979-314-8301
Education
- Undergraduate Education
- Pharm.D. University of Patras, Greece (1989)
- Graduate Education
- Ph.D. Tufts University (1994)
- Postdoc. Harvard Medical School, MGH Cancer Center (1999)
Areas of Expertise
- Molecular genetics
- RNA biology
- Metabolism
- Cell cycle
Professional Summary
Our goal is to understand how cells couple their growth and division. By learning how metabolism affects the process of cell division, we can control how quickly cells multiply. We are using baker’s yeast as a model organism to study this problem because it has several beneficial properties:
● The formation of a bud marks the start of cell division in yeast, allowing us to track the timing of cell division using a microscope.
● Yeast can grow in continuous cultures, so we can precisely control and monitor their metabolism.
● Yeast is a eukaryote that is easy to study genetically. Its cell division machinery is very similar to that of human cells.
Protein synthesis and cell division
The production of proteins is essential for cell growth and controls how quickly cells multiply. Understanding the role of protein synthesis in cell division will help us learn how new cell cycles begin, how fast they proceed, how many times they happen, and how the available nutrients control cell multiplication. These cellular decisions are often disrupted in diseases.
H. M. Blank, C. Reuse, K. Schmidt-Hohagen, S. E. Hammer, K. Hiller, M. Polymenis. Branched-chain amino acid synthesis is coupled to TOR activation early in the cell cycle in yeast. EMBO Rep. 24(9):e57372 (2023).
Translational control in ribosomal protein mutants
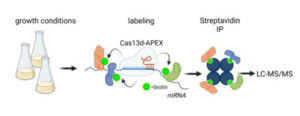
It is not well understood which proteins interact with specific mRNAs during the cell cycle. It is also easier to identify which mRNAs a specific protein might interact with than the other way around. We use a new CRISPR-based technology for RNA-centric proximity labeling to identify proteins interacting with particular RNAs during cell division. This technology allows us to identify all the proteins close to those RNAs. This will help us to understand how RNAs interact with proteins during cell division and how these interactions affect the cell cycle.
H. M. Blank, W. P. Griffith, M. Polymenis. Targeting APEX2 to the mRNA encoding fatty acid synthase β in yeast identifies interacting proteins that control its abundance in the cell cycle. Mol Biol Cell. 34(13):br20 (2023).
All Publications
- View publications on NIH