-
Wenshe Liu
- Harry E. Bovay Jr. Endowed Chair, Chemistry; Professor, Chemistry, Translational Medical Sciences, Biochemistry and Biophysics, Cell Biology and Genetics; Director, Texas A&M Drug Discovery Center
- Focus Area: COVID-19 drug discovery, chromatin biology, cancer epigenetics, posttranslational histone modifications, cancer drug discovery, the genetic code expansion, medicinal chemistry, structural biology, chemical biology
- Office:
- CHEM 204
- Email:
- [email protected]
- Phone:
- 979-845-1746
Education
- Undergraduate Education
- B.S. 2000
- Graduate Education
- Ph.D. 2005
- Postdoc. 2007
Areas of Expertise
- Chemical Biology
- Cancer Epigenetics
- Chemical Proteomics
- Drug Discovery
- Medicinal Chemistry
Professional Summary
We are a chemical biology research group with broad interests in fundamental biological processes, biotechnological innovation, and the development of novel therapeutics. The group specializes in techniques including the genetic code expansion technique that allows the formation of artificial organisms with noncanonical amino acids, phage display with unnatural peptide libraries, and the synthesis of small molecules, peptides, and proteins. Applications of these techniques include the understanding of epigenetic modifications on chromatin, the study of histone biology, phage-assisted drug discovery, and the development of small-molecule therapeutics.
COVID-19 drug discovery
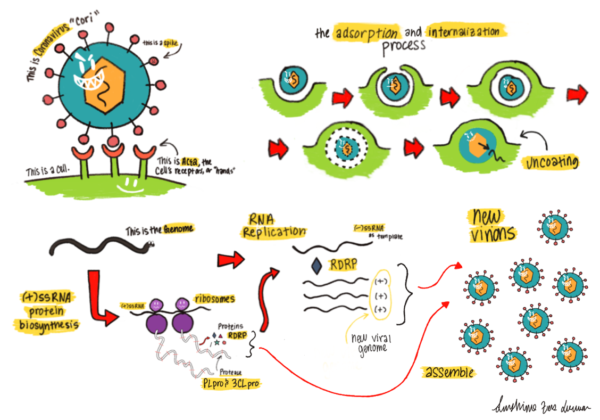
Lifecycle of a coronavirus entering and replicating inside a host cell. The (+)-stranded RNA is released upon viral entry, which starts the process of generating the viral coat and replicating the RNA genome.
See BioRxiv (2020).
SARS-CoV-2 is a newly emerged human coronavirus pathogen that has caused the COVID-19 pandemic. Rival only to the 1918 influenza pandemic, the COVID-19 pandemic has led to a profoundly catastrophic impact on the whole world. As of June 11, 2020, the total number of global COVID-19 cases has surpassed 7.4 million, of which more than 420,000 have succumbed to the deadly virus. By forcing many countries and regions into lockdown, COVID-19 is driving the world economy into a recession. To alleviate catastrophic damages of COVID-19 on public health, society, and economy, finding timely treatment options is of paramount importance. Since January 2020, the Liu group has been actively engaged in the identification of COVID-19 therapeutics. By targeting two essential enzymes of SARS-CoV-2, 3CLpro, and PLpro, both small molecules and peptide-based therapeutics have been developed. Some of these molecules have exhibited potency in inhibiting SARS-CoV-2 in infected cells at nanomolar levels.
E. C. Vatansever, K. Yang, K. C. Kratch, A. Drelich, C.-C. Cho, D. M. Mellott, S. Xu, C.-T. K. Tseng, W. R. Liu, Targeting the SARS-CoV-2 main protease to repurpose drugs for COVID-19. BioRxiv (2020).
The development of genetic code expansion technique
(A) A diagram to illustrate a chemical biology approach that probes sirtuin-targeted nucleosomal deacylation sites. (B) SIRT6 activities on 9 OcK-containing nucleosomes. EtBr stained the nucleosome DNA.
See J. Am. Chem. Soc. 141, 2472–2473 (2019).
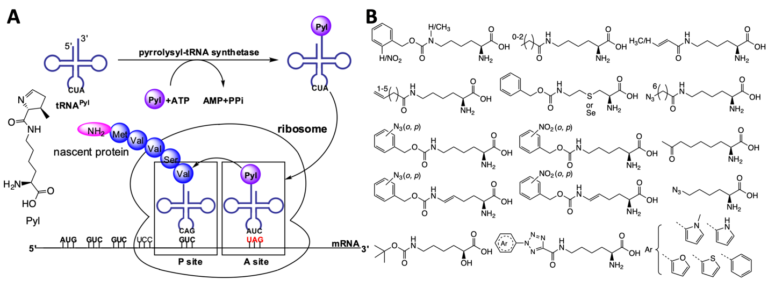
Pyrrolysine is the 22nd proteinogenic amino acid that was originally discovered in methanogenic archaea and some bacteria. Its coding involves amber codon, pyrrolysyl-tRNA synthetase (PylRS), amber suppressing tRNAPyl, and necessary enzymes for the synthesis of Pyl. Being a naturally occurring and well-evolved amber suppression system, the PylRS-tRNAPyl pair has turned into an enormous research tool for undergoing the amber suppression-based noncanonical amino acid (ncAA) mutagenesis in both bacteria and eukaryotic cells to produce functionally unique proteins. As one of the pioneers in the PylRS engineering research field, we have played an essential role in ascending the Pyl incorporation system as the most popular research tool for the ncAA mutagenesis. So far, nearly 200 ncAAs have been genetically encoded using native or evolved Pyl systems. More than 80 of them were developed in the Liu group. These ncAAs contain a large variety of functionalities with applications in both industry and academic research. We will continue our endeavor to develop novel genetic code expansion systems.
Z. A. Wang, Y. Kurra, X. Wang, Y. Zeng, Y. J. Lee, V. Sharma, H. Lin, S. Y. Dai, W. R. Liu, A versatile approach for site-specific lysine acylation in proteins. Angew. Chem. Int. Ed. 56, 1643–1647 (2017).
Understanding epigenetic functions of posttranslational histone modifications
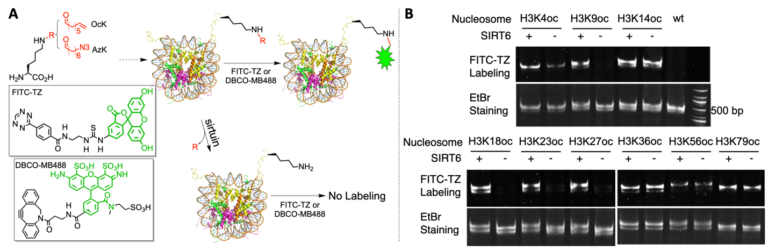
A) A diagram to illustrate a chemical biology approach that probes sirtuin-targeted nucleosomal deacylation sites. (B) SIRT6 activities on 9 OcK-containing nucleosomes. EtBr stained the nucleosome DNA. See J. Am. Chem. Soc. 141, 2472–2473 (2019). See Nat. Commun. 11 (2020).
Using the genetic code expansion technique, we have enabled histones’ synthesis with a large variety of posttranslational lysine modifications. The access of histones with defined modifications has been a major obstacle of understanding epigenetic functions of nucleosomal modifications during cellular development and differentiation. By using delicately designed and engineered cells, our team has demonstrated the synthesis of large quantities of histones with site-specific lysine monomethylation, acetylation, propionylation, butyrylation, and crotonylation. Moreover, we have devised a versatile method for the synthesis of histones potential with any lysine acylation. Using these approaches, we have demonstrated the constitution of nucleosomes site-specifically with lysine modifications in their histones and successfully applied these modified nucleosomes to investigate epigenetic eraser functions such as histone deacetylases. With our unique tools available, we are working toward the use of Cryo-EM to structurally characterize functions of lysine modifications in regulating the nucleosome structure and recognition of nucleosome modifications by epigenetic erasers and readers.
W. W. Wang, M. Angulo-Ibanez, J. Lyu, Y. Kurra, Z. Tong, B. Wu, L. Zhang, V. Sharma, J. Zhou, H. Lin, Y. Q. Gao, W. Li, K. F. Chua, W. R. Liu, A click chemistry approach reveals the chromatin-dependent histone H3K36 deacylase nature of SIRT7. J. Am. Chem. Soc. 141, 2472–2473 (2019).
Phage-displayed unnatural peptide libraries for drug discovery
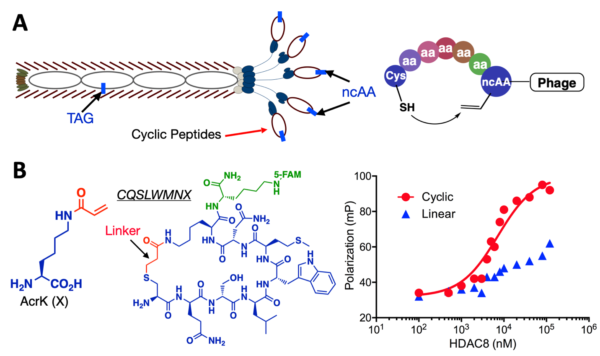
(A) An amber suppression-based approach to link a phenotypic ncAA with a genotypic TAG mutation and a proximity-driven cyclization reaction between a cysteine and an electrophilic ncAA in a phage-displayed peptide. (B) The structure of AcrK, an AcrK-based cyclic peptide inhibitor that was selected for HDAC8 and conjugated to a FITC fluorophore, and its binding to HDAC8. A linearized peptide was included for comparison. The binding was characterized using the fluorescence anisotropy assay.
See Nat. Commun. 11 (2020).
The production of M13 phages in E. coli, a popular host for the ncAA mutagenesis technique, makes it possible to merge the two techniques. Our exploration in this new and relatively applied research direction has led to the development of two novel methods, with one being the construction of genetically encoded, phage-displayed cyclic peptide libraries, and the other the active site-directed ligand evolution technique (ADLE) for phage display. In ADLE, a chemical warhead with a known capability to bind an active target site integrates into a ncAA that genetically incorporates into a phage-displayed sequence-randomized peptide library. During selection against the target, the binding of the ncAA to the target active site allows for a rationally designed interaction to guide the flanking residues in phage-displayed peptides toward the active site, increasing the probability that a given selection produces an effective, highly potent inhibitor. We are applying both techniques to identify inhibitors for both COVID-19 and cancer targets such as histone deacetylases.
J. M. Tharp, J. T. Hampton, C. A. Reed, A. Ehnbom, P. C. Chen, J. S. Morse, Y. Kurra, L. M. Perez, S. Xu, W. R. Liu, An amber obligate active site-directed ligand evolution technique for phage display. Nat. Commun. 11 (2020).