
-
Margaret Glasner
- Associate Professor, Biochemistry and Biophysics
- Focus Area: protein evolution, enzyme promiscuity, metabolic pathway evolution, metabolic pathway discovery, comparative genomics
- Office:
- BICH 237A
- Email:
- [email protected]
- Phone:
- 979-458-0123
Education
- Undergraduate Education
- B.S. University of Wyoming (1995)
- B.M. University of Wyoming (1995)
- Graduate Education
- Ph.D. Massachusetts Institute of Technology (2003)
- Postdoc. University of California, San Francisco (2003-2008)
Areas of Expertise
- Protein Evolution
- Enzymes
- Metabolism
Professional Summary
Evolution is the organizing principle of biology and provides the cornerstone of our approach to understand the relationships between protein structure and function. My laboratory employs a unique combination of molecular evolution, bioinformatics, and protein chemistry to investigate the evolution and biophysical basis of enzyme specificity. Using these tools, we generate a picture of enzyme evolution from the atomic level to the genomic assembly of metabolic pathways.
A major focus of the lab is to discover how catalytic promiscuity serves as the raw material for evolving new enzyme activities. Catalytic promiscuity is the ability to catalyze different chemical reactions using the same active site. We study this problem on two fronts: understanding the evolutionary potential of catalytically promiscuous proteins and discovering how enzymes that have promiscuous activities can be recruited to evolve new metabolic pathways. Another research focus capitalizes on catalytically promiscuous enzymes, along with experimental and bioinformatic analysis of functionally diverse enzyme superfamilies to investigate the biophysical basis of enzyme specificity.
Our overall goal is to use results from our research to identify fundamental evolutionary principles that can help decipher protein structure-function relationships, predict protein functions, and improve protein or metabolic engineering methods.
Mechanistic basis of catalytic promiscuity
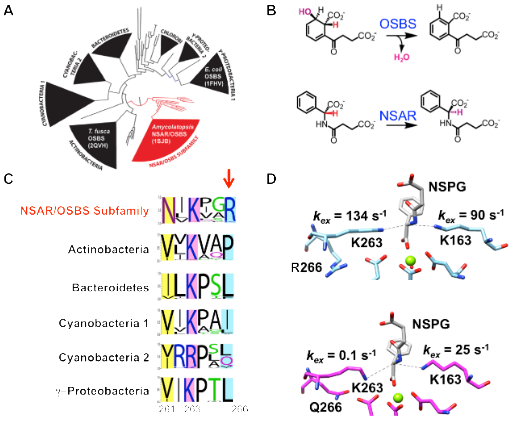
Figure 1: Identification of a specificity determinant in an NSAR/OSBS enzyme. A) The OSBS family can be divided into several structurally distinct subfamilies. NSAR activity evolved in one subfamily, shown in red. B) The OSBS and NSAR reactions use structurally similar substrates to catalyze different reactions. C) Sequence logos of each subfamily show that R266 is conserved in the NSAR/OSBS subfamily, while position 266 is primarily nonpolar in other OSBS subfamilies. D) Hydrogen-deuterium exchange experiments show that mutating the conserved R266 (top) to glutamine (bottom) severely reduces the rate of proton exchange between the catalytic K263 and the substrate. (Biochemistry 51, 6171–6181 (2012); Biochemistry 60, 3829-3840 (2021)).
We are investigating the mechanistic basis of promiscuity in enzymes that catalyze o-succinylbenzoate synthesis (OSBS) and N-succinylamino acid racemization (NSAR). NSAR activity originated as a promiscuous activity in one subfamily of the large, diverse OSBS enzyme family. Our results so far indicate that substrate orientation and reactivity of catalytic amino acids, rather than substrate-binding affinity, are the keys to catalytic promiscuity of NSAR/OSBS enzymes. Ancestral sequence reconstruction and structural comparisons identified a tyrosine to
isoleucine/leucine mutation, which modified the shape and size of the active site to enable N-succinylamino acids to bind in the right orientation for catalysis. Comparing sequence conservation in the NSAR/OSBS subfamily to other OSBS subfamilies, which lack NSAR activity, identified a second-shell arginine that is also essential for NSAR activity in Amycolatopsis sp. T-1-60 NSAR/OSBS. Mutating this arginine showed that it determines the reactivity of an adjacent catalytic lysine, a general acid-base catalyst in the NSAR reaction, but serves as a cation to stabilize the transition state in the OSBS reaction. We are continuing to search for and evaluate other mutations necessary for the evolution of NSAR activity using ancestral reconstruction, sequence/structure comparisons, and library screening approaches.
D. Odokonyero, A. W. McMillan, U. A. Ramagopal, R. Toro, D. P. Truong, M. Zhu, M. S. Lopez, B. Somiari, M. Herman, A. Aziz, J. B. Bonanno, K. G. Hull, S. K. Burley, D. Romo, S. C. Almo, M. E. Glasner, Comparison of Alicyclobacillus acidocaldarius o-succinylbenzoate synthase to its promiscuous N-succinylamino acid racemase/ o-succinylbenzoate synthase relatives. Biochemistry 57, 3676–3689 (2018). doi: 10.1021/acs.biochem.8b00088. PMID: 29767960; PMCID: PMC7187728.
Truong, D. P., Rousseau, S., Machala, B. W., Huddleston, J. P., Zhu, M., Hull, K. G., Romo, D., Raushel, F. M., Sacchettini, J. C., and Glasner, M. E. (2021) Second-Shell Amino Acid R266 Helps Determine N-Succinylamino Acid Racemase Reaction Specificity in Promiscuous N-Succinylamino Acid Racemase/o-Succinylbenzoate Synthase Enzymes, Biochemistry 60, 3829-3840. doi: 10.1021/acs.biochem.1c00627. PMID: 34845903; PMCID: PMC8939854
Biophysical basis of intramolecular epistasis
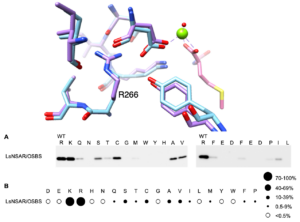
Figure 2: Despite sequence conservation of residues surrounding R266, mutating R266 to glutamine has a small effect on expression and OSBS activity of Amycolatopsis sp. T-1-60 NSAR/OSBS but is deleterious for the expression and both activities of Lysinibacillus sphaericus NSAR/OSBS. A) Structural superposition of Amycolatopsis sp. T-1-60 NSAR/OSBS (PDB: 1SJC, blue) and Lysinibacillus sphaericus NSAR/OSBS (Alphafold, purple). The magnesium ion (lime green) and N-succinylmethionine (salmon) are from PDB: 1SJC. B) Semi-quantitative western blot of Lysinibacillus sphaericus NSAR/OSBS variants with arginine (wild type) and other amino acids at position 266.
Intramolecular epistasis occurs when mutations have different effects in different sequence contexts; as a result, mutations within a protein have non-additive effects. Recent studies demonstrate that epistasis has a major influence on evolutionary trajectories of proteins. As a result, identifying epistatic interactions and understanding how they affect protein structure-function relationships is essential for understanding how proteins carry out their functions and engineering proteins. Catalytically promiscuous enzymes offer an unprecedented opportunity to investigate the biophysical basis of epistasis because epistatic mutations that affect stability and folding are likely to affect all the enzyme’s activities. In contrast, epistatic mutations that affect catalysis may alter the relative specificity among the activities. For example, in contrast to the preferential effect of R266Q on NSAR activity in Amycolatopsis sp. T-1-60 NSAR/OSBS, R266Q mutations in several other enzymes are highly deleterious for both NSAR and OSBS activities, indicating a broader role in protein conformation or stability. We are using computational and experimental methods to broadly map and characterize the effects of epistatic interactions within NSAR/OSBS enzymes. This will provide a comprehensive understanding of how epistatic interactions among residues influence protein structure-function relationships.
Truong, D. P., Fults, S., Davila, C., Huddleston, J. P., Brock, D., Zhu, M., Pellois, J.-P., Hull, K. G., Romo, D., Raushel, F. M., and Glasner, M. E. (2022) Roles of the second-shell amino acid R266 in other members of the MLE subgroup of the enolase superfamily, bioRxiv, 2022.2003.2003.481873 [Preprint]. March 481873, 482022 [cited 482022 Mar 481875].
Underground metabolism in the evolution of new metabolic pathways
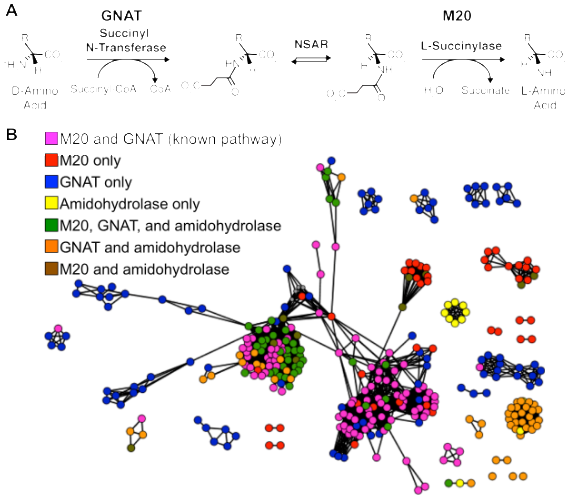
Figure 3: Comparative genomics shows that NSAR genes occur in a variety of operon contexts. A) Characterized pathway that utilizes NSAR activity (Biochemistry 45, 4455–4462 (2006)). B) Genome context network showing variations among the genes that are encoded next to an NSAR gene.
Metabolic pathways require the coordinated expression, substrate specificity, and kinetic properties of multiple enzymes. As a result, the evolution of new metabolic pathways is a complicated problem, requiring several low-probability events. Catalytic promiscuity and broad substrate specificity comprise an “underground metabolism” of side reactions that could be recruited to form new metabolic pathways. Events, such as mutations that alter flux through an underground reaction or horizontal gene transfer, could solidify connections among underground and normal metabolic reactions to form new metabolic pathways in a single step. We are investigating the evolution of new metabolic pathways that utilize NSAR activity to understand the combined contributions of horizontal gene transfer and underground metabolism. Our initial comparative genomics studies show that nsar genes, which have been horizontally transferred to many different microbial species, have been integrated into various genome contexts. This suggests that new pathways that utilize NSAR enzymes have evolved multiple times. We are using molecular phylogenetics to trace the origins of these metabolic pathways and experimentally characterizing the enzymes involved. Furthermore, we are using experimental evolution to investigate the de novo evolution of new metabolic pathways.
M. E. Glasner, D. P. Truong, B. C. Morse, How enzyme promiscuity and horizontal gene transfer contribute to metabolic innovation. FEBS J. 287, 1323–1342 (2020). doi: 10.1111/febs.15185. PMID: 31858709; PMCID: PMC7245361.
All Publications
- View publications on PubMed