
-
Jae-Hyun Cho
- Associate Professor, Biochemistry and Biophysics
- Focus Area: protein-protein interactions, NMR, crystallography, structural biology, protein motion, inhibitor development, influenza virus, thermodynamics, kinetics
- Office:
- NMR N113A
- Email:
- [email protected]
- Phone:
- 979-314-8307 Lab phone: 979-314-8308
Education
- Undergraduate Education
- B.S. Han-Yang University (1997)
- Graduate Education
- Ph.D. SUNY Stony Brook (2006)
- Postdoc. Columbia University (2012)
Areas of Expertise
- Molecular recognition between host and viruses
- Protein-protein/RNA interaction
- Protein dynamics and Structural biophysics
- NMR, Cryo-EM, and Crystallography
- Molecular evolution
Professional Summary
Our research interests lie in the interface between biology and the physical sciences. Protein-protein interactions are involved in virtually all biological processes. One research area in our laboratory is to understand the biophysical mechanisms of protein-protein interactions. Specifically, we investigate the molecular recognition mechanism between influenza viral proteins and human signaling proteins, and seek to elucidate the structural, thermodynamic, and kinetic mechanisms of the protein-protein interactions.
Protein-protein interactions between host and virus
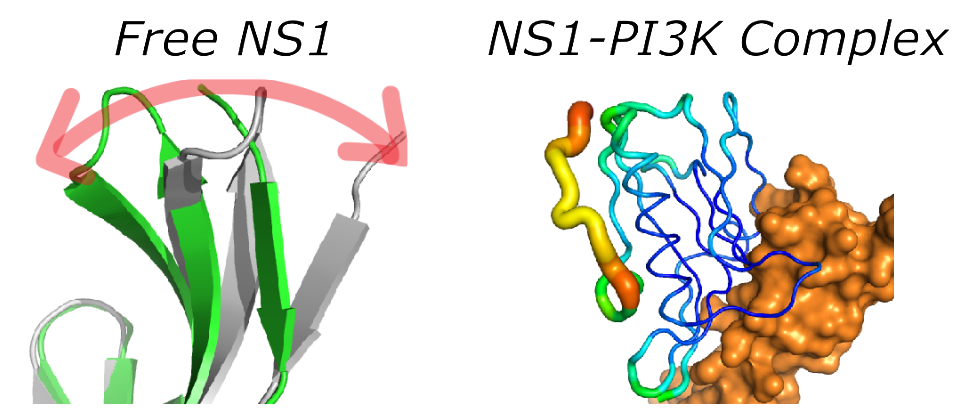
Role of protein motion in host-virus interaction. See PNAS 117, 6550–6558 (2020).
Influenza viruses continue to pose a serious pandemic threat. In the last century, there were four major flu pandemics. In particular, the 1918 influenza A virus (Spanish flu) was the most severe flu pandemic in human history. The protein NS1 is responsible for counteracting host innate immune responses during the infection cycle. Despite its importance, it is not well-understood how NS1 interacts with a number of host proteins. We are interested in how a viral virulence factor, nonstructural protein 1 (NS1), hijacks host signaling proteins. The current study in our lab employs NS1 from diverse influenza strains, including the 1918 virus. Using powerful structural biology techniques, such as NMR spectroscopy and X-ray crystallography, we investigate how the structure and conformational dynamics of NS1 play a role in the interaction with host proteins.
J.-H. Cho, B. Zhao, J. Shi, N. Savage, Q. Shen, J. Byrnes, L. Yang, W. Hwang, P. Li, Molecular recognition of a host protein by NS1 of pandemic and seasonal influenza A viruses. Proc. Natl. Acad. Sci. 117, 6550–6558 (2020).

Ternary interaction of NS1 with host PI3K and CRK.
See Viruses 12, 338–348 (2020).
Viral proteins evade host innate immune responses
The host innate immune response plays a critical role as a front-line defense mechanism against viral infection. Influenza viruses use NS1 to evade or counteracts the innate immune responses. However, many questions remain to be addressed as to the molecular basis underlying how NS1 blocks the immune responses. Moreover, it is unclear whether NS1 of different influenza strains has a distinct mechanism to counteract the immune responses. We aim to reveal the detailed structural basis of the interaction between NS1 and host proteins responsible for innate immune responses.
A. Dubrow, L. Sirong, N. Savage, Q. Shen, J.-H. Cho, Molecular basis of the ternary interaction between NS1 of the 1918 influenza A virus, PI3K, and CRK. Viruses 12, 338–348 (2020).
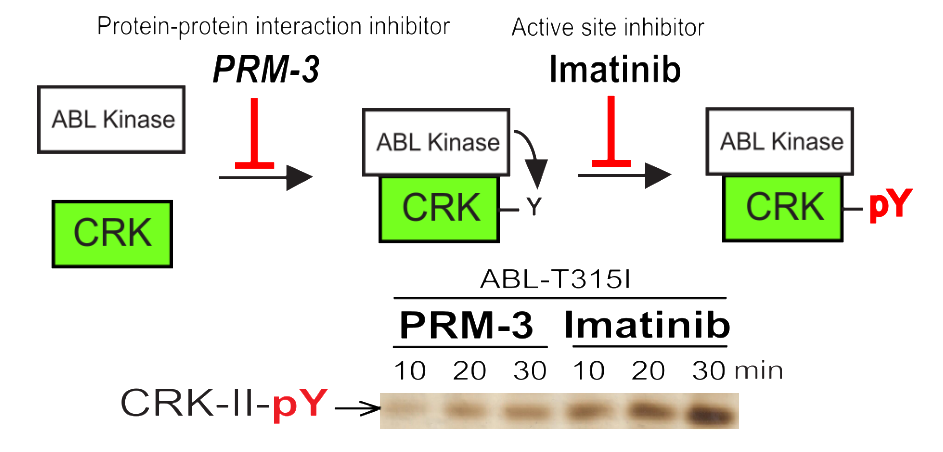
Protein-protein interaction inhibitors. See Med. Chem. Comm. 9, 519–524 (2018).
Development of protein-protein interaction inhibitors
Another research area is to develop protein-protein interaction inhibitors, where we actively integrate chemical biology approaches with biophysical approaches. We also seek to develop a new technology that can be applied to so-called “undruggable” proteins. We currently focus on how to block oncogenic protein-protein interactions and viral hijacking of host proteins.
Q. Shen, J. Shi, D. Zeng, B. Zhao, P. Li, W. Hwang, J.-H. Cho. Molecular mechanisms of tight binding through fuzzy interactions. Biophys. J. 114 ,1313–1320 (2018).
All Publications
- View publications on NIH