
-
Mary Bryk
- Associate Dean for Academic Affairs, College of Agriculture and Life Sciences and Associate Professor, Biochemistry & Biophysics, Genetics
- Focus Area: transcriptional regulation, gene expression, chromatin, histone modifications, molecular genetics (NOT CURRENTLY TAKING GRADUATE STUDENTS)
- Office:
- BICH 234
- Email:
- [email protected]
- Phone:
- 979-862-2294
Education
- Undergraduate Education
- B.S. Cornell University (1985)
- Graduate Education
- Ph.D. Albany Medical College (1994)
- Postdoc. NY State Department of Health, David Axelrod Institute (1994-1997)
- Postdoc. Harvard Medical School (1997-2002)
- Courses Taught
- BICH/GENE 673 GENE EXPRESSION
Areas of Expertise
- Transcriptional regulation
- Gene expression
- Chromatin
- Histone modifications
- Molecular genetics
Professional Summary
How cells regulate the expression of genes is fundamental to most aspects of biology. My laboratory is interested in understanding the regulation of gene expression at active and inactive regions of the yeast genome. The goal is to learn about mechanisms that regulate Pol II transcription and chromatin structure. We identify proteins that regulate transcription using classic and modern genetic techniques. We then investigate the mechanisms through which these factors influence transcription and chromatin function using a combination of genetic, molecular, and biochemical methods. These methods include mutagenesis, genetic screens, growth assays, RNA-seq, and chromatin immunoprecipitation. Our findings will explain how chromatin dynamics influence gene expression and genome integrity. Many of the factors we study have homologs in human cells, indicating that our discoveries may provide insights into mechanisms that regulate gene expression in higher eukaryotes.
Methylation of histone H3 by Set1 regulates transcription
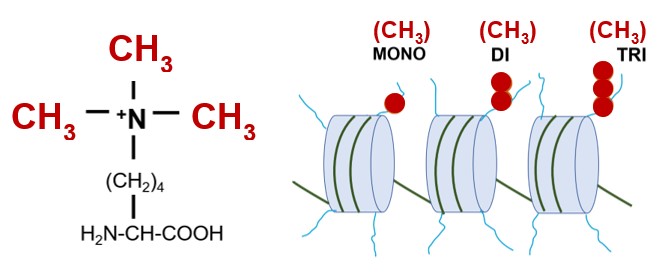
Set1 is a lysine methyltransferase that catalyzes the methylation of a lysine residue (K4) in the N terminal tail of histone H3 proteins. Histone H3 proteins are subunits of nucleosomes that comprise eukaryotic chromatin. Left, a trimethylated lysine residue; right, chromatin with nucleosomes with histone H3 proteins that are mono, di or trimethylated on K4 by Set1.
Our current research addresses complex questions regarding the role of histone methylation in the regulation of transcription. In budding yeast, there is a single lysine methyltransferase, Set1, that catalyzes the mono-, di- and tri-methylation of the fourth residue, lysine 4 (K4), of histone H3. The SET family of lysine methyltransferases is evolutionarily conserved in structure and function. Set1 is a great model to decipher the effects of histone methylation on gene expression and chromatin function because it is the only histone H3K4 specific methyltransferase in S. cerevisiae. Set1 generates three chromatin marks, and each mark is not equivalent to gene regulation. We exploit novel genetic variants of the conserved Set1 protein that differentially affect methylation of lysine 4 of histone H3 (e.g., abolish H3K4 tri-methylation while keeping H3K4 mono-methylation intact). Studies using these Set1 mutants provide insights into the roles of different H3K4 methyl marks in transcription by Pol II. This research is expected to define new paradigms in gene regulation.
K. Williamson, V. Schneider, R. A. Jordan, M. Henderson Pozzi, J. E. Mueller, M. Bryk, Catalytic and functional roles of conserved amino acids in the SET domain of the S. cerevisiae lysine methyltransferase Set1. PLoS One 8 (2013).
N. Deshpande, R. Jordan, M. Henderson-Pozzi, M. Bryk, Histone 3 lysine 4 monomethylation supports activation of transcription in S. cerevisiae during nutrient stress. Curr Genet 68 181-194 (2022)
N. Deshpande, M. Bryk, Diverse and dynamic forms of gene regulation by the S. cerevisiae histone methyltransferase Set1. Curr Genet 69 91-114 (2023)
All Publications
- View publications on Google Scholar